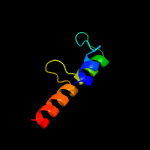
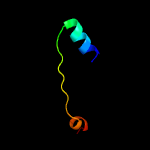
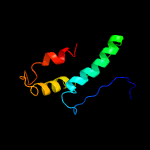
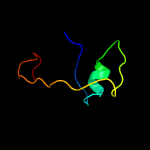
1 c2rkhA_
51.2
36
PDB header: transcriptionChain: A: PDB Molecule: putative apha-like transcription factor;PDBTitle: crystal structure of a putative apha-like transcription factor2 (zp_00208345.1) from magnetospirillum magnetotacticum ms-1 at 2.00 a3 resolution
2 c4dt1B_
30.3
36
PDB header: dna binding proteinChain: B: PDB Molecule: platinum sensitivity protein 3;PDBTitle: crystal structure of the psy3-csm2 complex
3 c2v6xB_
28.5
50
PDB header: protein transportChain: B: PDB Molecule: doa4-independent degradation protein 4;PDBTitle: stractural insight into the interaction between escrt-iii and vps4
4 d2pbka1
22.2
35
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin
5 c3njqB_
21.7
29
PDB header: viral protein/inhibitorChain: B: PDB Molecule: orf 17;PDBTitle: crystal structure of kaposi's sarcoma-associated herpesvirus protease2 in complex with dimer disruptor
6 d1at3a_
21.3
46
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin
7 d2cyja1
19.8
22
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
8 c2jqmA_
19.0
33
PDB header: transferaseChain: A: PDB Molecule: envelope protein e;PDBTitle: yellow fever envelope protein domain iii nmr structure2 (s288-k398)
9 c3hjeA_
18.3
32
PDB header: transferaseChain: A: PDB Molecule: 704aa long hypothetical glycosyltransferase;PDBTitle: crystal structure of sulfolobus tokodaii hypothetical maltooligosyl2 trehalose synthase
10 c3egpA_
17.7
28
PDB header: viral proteinChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure analysis of dengue-1 envelope protein2 domain iii
11 d2al3a1
17.6
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain
12 d1iega_
16.3
22
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin
13 c1fx0B_
15.9
19
PDB header: hydrolaseChain: B: PDB Molecule: atp synthase beta chain;PDBTitle: crystal structure of the chloroplast f1-atpase from spinach
14 c2vdcI_
15.9
24
PDB header: oxidoreductaseChain: I: PDB Molecule: glutamate synthase [nadph] small chain;PDBTitle: the 9.5 a resolution structure of glutamate synthase from cryo-2 electron microscopy and its oligomerization behavior in solution:3 functional implications.
15 c2h0pA_
15.5
22
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: nmr structure of the dengue-4 virus envelope protein domain2 iii
16 d1ok8a1
14.4
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain
17 d2fmla2
12.4
20
Fold: NudixSuperfamily: NudixFamily: BT0354 N-terminal domain-like
18 c3nrhA_
12.0
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bf1032;PDBTitle: crystal structure of protein bf1032 from bacteroides fragilis,2 northeast structural genomics consortium target bfr309
19 d1s6na_
11.3
42
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain
20 d2o3aa1
11.1
29
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF0751-like
21 d2fvta1
not modelled
11.0
18
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
22 c3uajA_
not modelled
10.9
22
PDB header: viral protein/immune systemChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure of the envelope glycoprotein ectodomain from dengue2 virus serotype 4 in complex with the fab fragment of the chimpanzee3 monoclonal antibody 5h2
23 d2fi9a1
not modelled
10.8
33
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
24 c2yrcA_
not modelled
10.7
71
PDB header: protein transportChain: A: PDB Molecule: protein transport protein sec23a;PDBTitle: solution structure of the zf-sec23_sec24 from human sec23a
25 c6epkA_
not modelled
10.7
33
PDB header: viral proteinChain: A: PDB Molecule: envelope protein e;PDBTitle: crystal structure of the precursor membrane protein-envelope protein2 heterodimer from the yellow fever virus
26 c3fz5C_
not modelled
10.3
24
PDB header: isomeraseChain: C: PDB Molecule: possible 2-hydroxychromene-2-carboxylate isomerase;PDBTitle: crystal structure of possible 2-hydroxychromene-2-carboxylate2 isomerase from rhodobacter sphaeroides
27 c4ip8B_
not modelled
10.3
30
PDB header: protein bindingChain: B: PDB Molecule: serum amyloid a-1 protein;PDBTitle: structure of human serum amyloid a1
28 d1fx0a3
not modelled
10.0
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)
29 d1o6ea_
not modelled
9.9
25
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin
30 c2k2eA_
not modelled
9.8
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bp2786;PDBTitle: solution nmr structure of bordetella pertussis protein2 bp2786, a mth938-like domain. northeast structural3 genomics consortium target ber31
31 c5x51X_
not modelled
9.7
38
PDB header: transferaseChain: X: PDB Molecule: rna polymerase subunit, found in rna polymerase complexesPDBTitle: rna polymerase ii from komagataella pastoris (type-3 crystal)
32 c2h6oA_
not modelled
9.6
19
PDB header: viral proteinChain: A: PDB Molecule: major outer envelope glycoprotein gp350;PDBTitle: epstein barr virus major envelope glycoprotein
33 c1uzgA_
not modelled
9.5
25
PDB header: viral proteinChain: A: PDB Molecule: major envelope protein e;PDBTitle: crystal structure of the dengue type 3 virus envelope2 protein
34 c2yxyA_
not modelled
9.3
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical conserved protein, gk0453;PDBTitle: crystarl structure of hypothetical conserved protein, gk0453
35 c5zcrB_
not modelled
9.3
38
PDB header: hydrolaseChain: B: PDB Molecule: maltooligosyl trehalose synthase;PDBTitle: dsm5389 glycosyltrehalose synthase
36 d1dqna_
not modelled
9.3
50
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
37 c2yy8B_
not modelled
9.1
29
PDB header: transferaseChain: B: PDB Molecule: upf0106 protein ph0461;PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
38 d1ztxe1
not modelled
9.0
42
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain
39 c6rdtY_
not modelled
9.0
23
PDB header: proton transportChain: Y: PDB Molecule: atp synthase subunit beta;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 composite map
40 c4xvwK_
not modelled
8.9
16
PDB header: isomeraseChain: K: PDB Molecule: dsba-like protein;PDBTitle: crystal structure of proteus mirabilis scsc in a compact conformation
41 d2iida1
not modelled
8.5
20
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
42 c3c6dB_
not modelled
8.4
22
PDB header: virusChain: B: PDB Molecule: polyprotein;PDBTitle: the pseudo-atomic structure of dengue immature virus
43 d1pjwa_
not modelled
8.2
39
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain
44 d1z6ma1
not modelled
8.2
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like
45 d2q4qa1
not modelled
7.9
22
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
46 c5whaF_
not modelled
7.6
68
PDB header: protein bindingChain: F: PDB Molecule: miniprotein 225-11;PDBTitle: kras g12v, bound to gdp and miniprotein 225-11
47 c1hznA_
not modelled
7.6
45
PDB header: hormone/growth factorChain: A: PDB Molecule: cholecystokinin type a receptor;PDBTitle: nmr solution structure of the third extracellular loop of2 the cholecystokinin a receptor
48 d1na6a2
not modelled
7.6
19
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Type II restriction endonuclease catalytic domain
49 d2ifxa1
not modelled
7.5
39
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: MmlI-like
50 c6eh1D_
not modelled
7.5
70
PDB header: virusChain: D: PDB Molecule: minor capsid protein micp;PDBTitle: sacbrood virus of honeybee - expansion state ii
51 c6egvD_
not modelled
7.5
70
PDB header: virusChain: D: PDB Molecule: minor capsid protein micp;PDBTitle: sacbrood virus of honeybee
52 c6egxD_
not modelled
7.5
70
PDB header: virusChain: D: PDB Molecule: minor capsid protein micp;PDBTitle: sacbrood virus of honeybee - expansion state i
53 c5oypD_
not modelled
7.5
70
PDB header: virusChain: D: PDB Molecule: minor capsid protein micp;PDBTitle: sacbrood virus of honeybee
54 c5lsfD_
not modelled
7.5
70
PDB header: virusChain: D: PDB Molecule: vp4;PDBTitle: sacbrood honeybee virus
55 c2ja6L_
not modelled
7.1
38
PDB header: transferaseChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iii 7.7 kdaPDBTitle: cpd lesion containing rna polymerase ii elongation complex b
56 c6f5dE_
not modelled
7.1
21
PDB header: hydrolaseChain: E: PDB Molecule: atp synthase subunit beta, mitochondrial;PDBTitle: trypanosoma brucei f1-atpase
57 c4gxzB_
not modelled
7.1
16
PDB header: isomeraseChain: B: PDB Molecule: suppression of copper sensitivity protein;PDBTitle: crystal structure of a periplasmic thioredoxin-like protein from2 salmonella enterica serovar typhimurium
58 c2gm2A_
not modelled
7.1
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
59 c5flmL_
not modelled
6.9
31
PDB header: transcriptionChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iii subunit rpabc4;PDBTitle: structure of transcribing mammalian rna polymerase ii
60 c5j8tA_
not modelled
6.9
0
PDB header: hydrolaseChain: A: PDB Molecule: choline binding protein;PDBTitle: nmr structure of excalibur domain of cbpl
61 c2yewB_
not modelled
6.9
25
PDB header: virusChain: B: PDB Molecule: e1 envelope glycoprotein;PDBTitle: modeling barmah forest virus structural proteins
62 c3nuhB_
not modelled
6.7
57
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
63 d2alaa2
not modelled
6.6
63
Fold: Viral glycoprotein, central and dimerisation domainsSuperfamily: Viral glycoprotein, central and dimerisation domainsFamily: Viral glycoprotein, central and dimerisation domains
64 c4exnF_
not modelled
6.4
62
PDB header: cytokineChain: F: PDB Molecule: interleukin-34;PDBTitle: crystal structure of mouse interleukin-34
65 c6eezC_
not modelled
6.4
22
PDB header: isomeraseChain: C: PDB Molecule: dsba-like disulfide oxidoreductase;PDBTitle: crystal structure of the thiol-disulfide exchange protein alpha-dsba22 from wolbachia pipientis
66 c1kmhA_
not modelled
6.2
11
PDB header: hydrolaseChain: A: PDB Molecule: atpase alpha subunit;PDBTitle: crystal structure of spinach chloroplast f1-atpase complexed with2 tentoxin
67 c5d50E_
not modelled
6.2
42
PDB header: dna binding proteinChain: E: PDB Molecule: anti-repressor protein;PDBTitle: crystal structure of rep-ant complex from salmonella-temperate phage
68 c1na6B_
not modelled
6.2
15
PDB header: hydrolaseChain: B: PDB Molecule: restriction endonuclease ecorii;PDBTitle: crystal structure of restriction endonuclease ecorii mutant2 r88a
69 d1u0bb1
not modelled
6.0
32
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
70 d1t1ua2
not modelled
5.9
14
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase
71 c3h0gL_
not modelled
5.9
33
PDB header: transcriptionChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iii subunit rpabc4;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
72 d1iv8a2
not modelled
5.6
50
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
73 c6o55B_
not modelled
5.6
17
PDB header: isomeraseChain: B: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 (pure) from legionella pneumophila
74 c2zkr9_
not modelled
5.6
38
PDB header: ribosomal protein/rnaChain: 9: PDB Molecule: 60s ribosomal protein l32;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
75 d1vaea_
not modelled
5.4
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain
76 d3etja1
not modelled
5.4
43
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like
77 c6a5eD_
not modelled
5.4
21
PDB header: transferaseChain: D: PDB Molecule: gpi-anchored protein llg2;PDBTitle: crystal structure of plant peptide ralf23 in complex with fer and llg2
78 d1alna2
not modelled
5.3
20
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
79 c1l6jA_
not modelled
5.3
14
PDB header: hydrolaseChain: A: PDB Molecule: matrix metalloproteinase-9;PDBTitle: crystal structure of human matrix metalloproteinase mmp92 (gelatinase b).
80 c2of6C_
not modelled
5.3
42
PDB header: virusChain: C: PDB Molecule: envelope glycoprotein e;PDBTitle: structure of immature west nile virus
81 c4lh9A_
not modelled
5.2
20
PDB header: transcriptionChain: A: PDB Molecule: heterocyst differentiation control protein;PDBTitle: crystal structure of the refolded hood domain (asp256-gly295) of hetr
82 c1d6gA_
not modelled
5.2
55
PDB header: hormone/growth factorChain: A: PDB Molecule: cholecystokinin type a receptor;PDBTitle: molecular complex of cholecystokinin-8 and n-terminus of2 the cholecystokinin a receptor by nmr spectroscopy
83 c4k90A_
not modelled
5.2
46
PDB header: hydrolaseChain: A: PDB Molecule: extracellular metalloproteinase mep;PDBTitle: extracellular metalloproteinase from aspergillus
84 c2xznQ_
not modelled
5.1
27
PDB header: ribosomeChain: Q: PDB Molecule: ribosomal protein s17 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
85 c4cbfE_
not modelled
5.1
25
PDB header: virusChain: E: PDB Molecule: envelope protein e;PDBTitle: near-atomic resolution cryo-em structure of dengue serotype 4 virus