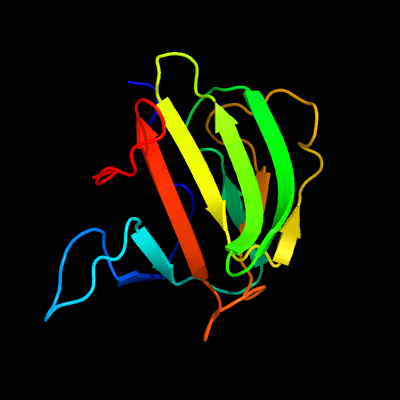
| 1 | c3sluB_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:m23 peptidase domain protein;
PDBTitle: crystal structure of nmb0315
|
|
|
|
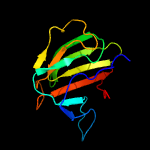
| 2 | c2gu1A_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:zinc peptidase;
PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
|
|
|
|
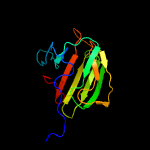
| 3 | c2hsiB_
|
|
 |
100.0 |
26 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative peptidase m23;
PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
|
|
|
|
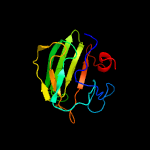
| 4 | c4rnzA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:conserved hypothetical secreted protein;
PDBTitle: structure of helicobacter pylori csd3 from the hexagonal crystal
|
|
|
|
| 5 | d1qwya_
|
|
 |
100.0 |
26 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Peptidoglycan hydrolase LytM |
|
|
|
| 6 | c4lxcA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysostaphin;
PDBTitle: the antimicrobial peptidase lysostaphin from staphylococcus simulans
|
|
|
|
| 7 | c3nyyA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative glycyl-glycine endopeptidase lytm;
PDBTitle: crystal structure of a putative glycyl-glycine endopeptidase lytm2 (rumgna_02482) from ruminococcus gnavus atcc 29149 at 1.60 a3 resolution
|
|
|
|
| 8 | c5j1mD_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:toxr-activated gene (tage);
PDBTitle: crystal structure of csd1-csd2 dimer ii
|
|
|
|
| 9 | c4bh5B_
|
|
 |
100.0 |
21 |
PDB header:cell cycle
Chain: B: PDB Molecule:murein hydrolase activator envc;
PDBTitle: lytm domain of envc, an activator of cell wall amidases in2 escherichia coli
|
|
|
|
| 10 | c5kvpA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:zoocin a endopeptidase;
PDBTitle: solution structure of the catalytic domain of zoocin a
|
|
|
|
| 11 | c5kqbA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase m23;
PDBTitle: identification and structural characterization of lytu
|
|
|
|
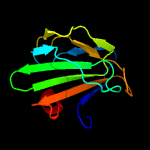
| 12 | c5j1lA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxr-activated gene (tage);
PDBTitle: crystal structure of csd1-csd2 dimer i
|
|
|
|
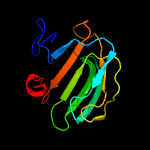
| 13 | c2b44A_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycyl-glycine endopeptidase lytm;
PDBTitle: truncated s. aureus lytm, p 32 2 1 crystal form
|
|
|
|
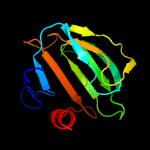
| 14 | c4qpbB_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:lysostaphin;
PDBTitle: catalytic domain of the antimicrobial peptidase lysostaphin from2 staphylococcus simulans crystallized in the absence of phosphate
|
|
|
|
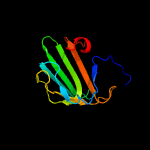
| 15 | c3tufB_
|
|
 |
100.0 |
22 |
PDB header:signaling protein
Chain: B: PDB Molecule:stage ii sporulation protein q;
PDBTitle: structure of the spoiiq-spoiiiah pore forming complex.
|
|
|
|
| 16 | c3uz0D_
|
|
 |
100.0 |
21 |
PDB header:transport protein
Chain: D: PDB Molecule:stage ii sporulation protein q;
PDBTitle: crystal structure of spoiiiah and spoiiq complex
|
|
|
|
| 17 | c5gt1A_
|
|
 |
100.0 |
25 |
PDB header:choline-binding protein
Chain: A: PDB Molecule:choline binding protein a;
PDBTitle: crystal structure of cbpa from l. salivarius ren
|
|
|
|
| 18 | c3it5B_
|
|
 |
99.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:protease lasa;
PDBTitle: crystal structure of the lasa virulence factor from pseudomonas2 aeruginosa
|
|
|
|
| 19 | c5b0hB_
|
|
 |
99.9 |
19 |
PDB header:metal binding protein
Chain: B: PDB Molecule:leukocyte cell-derived chemotaxin-2;
PDBTitle: crystal structure of human leukocyte cell-derived chemotaxin 2
|
|
|
|
| 20 | c5tz8C_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:glycosyl transferase;
PDBTitle: crystal structure of s. aureus tars
|
|
|
|
| 21 | c3f1yC_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: mannosyl-3-phosphoglycerate synthase from rubrobacter xylanophilus
|
|
|
| 22 | c3ckvA_ |
|
not modelled |
99.9 |
26 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a mycobacterial protein
|
|
|
| 23 | c3csqC_ |
|
not modelled |
99.9 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:morphogenesis protein 1;
PDBTitle: crystal and cryoem structural studies of a cell wall2 degrading enzyme in the bacteriophage phi29 tail
|
|
|
| 24 | c5mm1A_ |
|
not modelled |
99.9 |
18 |
PDB header:membrane protein
Chain: A: PDB Molecule:dolichol monophosphate mannose synthase;
PDBTitle: dolichyl phosphate mannose synthase in complex with gdp and dolichyl2 phosphate mannose
|
|
|
| 25 | d1omza_ |
|
not modelled |
99.9 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Exostosin |
|
|
| 26 | c5ekeB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:uncharacterized glycosyltransferase sll0501;
PDBTitle: structure of the polyisoprenyl-phosphate glycosyltransferase gtrb2 (f215a mutant)
|
|
|
| 27 | c2z86D_ |
|
not modelled |
99.9 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:chondroitin synthase;
PDBTitle: crystal structure of chondroitin polymerase from escherichia coli2 strain k4 (k4cp) complexed with udp-glcua and udp
|
|
|
| 28 | d1xhba2 |
|
not modelled |
99.9 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Polypeptide N-acetylgalactosaminyltransferase 1, N-terminal domain |
|
|
| 29 | c6h4mA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:probable ss-1,3-n-acetylglucosaminyltransferase;
PDBTitle: tarp-udp-glcnac-3rbop
|
|
|
| 30 | c1omxB_ |
|
not modelled |
99.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:alpha-1,4-n-acetylhexosaminyltransferase extl2;
PDBTitle: crystal structure of mouse alpha-1,4-n-acetylhexosaminyltransferase2 (extl2)
|
|
|
| 31 | c2ffuA_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 2;
PDBTitle: crystal structure of human ppgalnact-2 complexed with udp and ea2
|
|
|
| 32 | c1xhbA_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 1;
PDBTitle: the crystal structure of udp-galnac: polypeptide alpha-n-2 acetylgalactosaminyltransferase-t1
|
|
|
| 33 | c2d7iA_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 10;
PDBTitle: crystal structure of pp-galnac-t10 with udp, galnac and mn2+
|
|
|
| 34 | c6iwqE_ |
|
not modelled |
99.8 |
19 |
PDB header:transferase
Chain: E: PDB Molecule:n-acetylgalactosaminyltransferase 7;
PDBTitle: crystal structure of galnac-t7 with mn2+
|
|
|
| 35 | d1qg8a_ |
|
not modelled |
99.8 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Spore coat polysaccharide biosynthesis protein SpsA |
|
|
| 36 | c6e4rB_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 9;
PDBTitle: crystal structure of the drosophila melanogaster polypeptide n-2 acetylgalactosaminyl transferase pgant9b
|
|
|
| 37 | c5nqaA_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 4;
PDBTitle: crystal structure of galnac-t4 in complex with the monoglycopeptide 3
|
|
|
| 38 | c4fixA_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:udp-galactofuranosyl transferase glft2;
PDBTitle: crystal structure of glft2
|
|
|
| 39 | c5heaA_ |
|
not modelled |
99.8 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase (galt1);
PDBTitle: cgt structure in hexamer
|
|
|
| 40 | c3bcvA_ |
|
not modelled |
99.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase protein;
PDBTitle: crystal structure of a putative glycosyltransferase from bacteroides2 fragilis
|
|
|
| 41 | c5z8bB_ |
|
not modelled |
99.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:kfia protein;
PDBTitle: truncated n-acetylglucosaminyl transferase kfia from e. coli k5 strain2 apo form
|
|
|
| 42 | c4hg6A_ |
|
not modelled |
99.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:cellulose synthase subunit a;
PDBTitle: structure of a cellulose synthase - cellulose translocation2 intermediate
|
|
|
| 43 | c3zf8A_ |
|
not modelled |
99.6 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:mannan polymerase complexes subunit mnn9;
PDBTitle: crystal structure of saccharomyces cerevisiae mnn9 in2 complex with gdp and mn.
|
|
|
| 44 | c2qgiA_ |
|
not modelled |
99.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:atp synthase subunits region orf 6;
PDBTitle: the udp complex structure of the sixth gene product of the f1-atpase2 operon of rhodobacter blasticus
|
|
|
| 45 | c5ggfC_ |
|
not modelled |
99.4 |
18 |
PDB header:transferase, sugar binding protein
Chain: C: PDB Molecule:protein o-linked-mannose beta-1,2-n-
PDBTitle: crystal structure of human protein o-mannose beta-1,2-n-2 acetylglucosaminyltransferase form ii
|
|
|
| 46 | c6p61D_ |
|
not modelled |
99.4 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:glycosyltransferase;
PDBTitle: structure of a glycosyltransferase from leptospira borgpetersenii2 serovar hardjo-bovis (strain jb197)
|
|
|
| 47 | d2bo4a1 |
|
not modelled |
98.9 |
11 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:MGS-like |
|
|
| 48 | c6fxyA_ |
|
not modelled |
98.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:procollagen-lysine,2-oxoglutarate 5-dioxygenase 3;
PDBTitle: crystal structure of full-length human lysyl hydroxylase lh3 -2 cocrystal with fe2+, mn2+, udp-gal - structure from long-wavelength3 s-sad
|
|
|
| 49 | d1fo8a_ |
|
not modelled |
97.7 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:N-acetylglucosaminyltransferase I |
|
|
| 50 | c4irqB_ |
|
not modelled |
97.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:beta-1,4-galactosyltransferase 7;
PDBTitle: crystal structure of catalytic domain of human beta1,2 4galactosyltransferase 7 in closed conformation in complex with3 manganese and udp
|
|
|
| 51 | c5vcmA_ |
|
not modelled |
97.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-1,6-mannosyl-glycoprotein 2-beta-n-
PDBTitle: alpha-1,6-mannosyl-glycoprotein 2-beta-n-acetylglucosaminyltransferase2 with bound udp and manganese
|
|
|
| 52 | c2px7A_ |
|
not modelled |
96.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from thermus thermophilus hb8
|
|
|
| 53 | d2gpra_ |
|
not modelled |
96.3 |
19 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 54 | d2f3ga_ |
|
not modelled |
96.2 |
18 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 55 | d1glaf_ |
|
not modelled |
96.2 |
18 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 56 | d1gpra_ |
|
not modelled |
96.0 |
16 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 57 | d1pzta_ |
|
not modelled |
95.9 |
21 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:beta 1,4 galactosyltransferase (b4GalT1) |
|
|
| 58 | c2lmcB_ |
|
not modelled |
95.9 |
24 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: structure of t7 transcription factor gp2-e. coli rnap jaw domain2 complex
|
|
|
| 59 | d1vpaa_ |
|
not modelled |
95.7 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 60 | c2wvmA_ |
|
not modelled |
95.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: h309a mutant of mannosyl-3-phosphoglycerate synthase from2 thermus thermophilus hb27 in complex with3 gdp-alpha-d-mannose and mg(ii)
|
|
|
| 61 | c3okrC_ |
|
not modelled |
95.6 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
|
|
| 62 | d1vh3a_ |
|
not modelled |
95.5 |
7 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 63 | c2wawA_ |
|
not modelled |
95.3 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:moba relate protein;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0371c2 homolog from mycobacterium sp. strain jc1
|
|
|
| 64 | d1ci3m2 |
|
not modelled |
95.2 |
31 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
|
|
| 65 | c5ddtA_ |
|
not modelled |
95.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ispd from bacillus subtilis at 1.80 angstroms2 resolution, crystal form i
|
|
|
| 66 | d1vgwa_ |
|
not modelled |
94.8 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 67 | c2zu8A_ |
|
not modelled |
94.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: crystal structure of mannosyl-3-phosphoglycerate synthase2 from pyrococcus horikoshii
|
|
|
| 68 | c1w57A_ |
|
not modelled |
94.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:ispd/ispf bifunctional enzyme;
PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
|
|
|
| 69 | d1i52a_ |
|
not modelled |
94.4 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 70 | d1e2wa2 |
|
not modelled |
94.1 |
19 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
|
|
| 71 | c2aukA_ |
|
not modelled |
93.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of e. coli rna polymerase beta' g/g' insert
|
|
|
| 72 | c4kt7A_ |
|
not modelled |
93.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: the crystal structure of 4-diphosphocytidyl-2c-methyl-d-2 erythritolsynthase from anaerococcus prevotii dsm 20548
|
|
|
| 73 | c6oewB_ |
|
not modelled |
93.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:cytidylyltransferase;
PDBTitle: structure of a cytidylyltransferase from leptospira borgpetersenii2 serovar hardjo-bovis (strain jb197)
|
|
|
| 74 | c4ys8B_ |
|
not modelled |
93.2 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase (ispd) from burkholderia thailandensis
|
|
|
| 75 | c1e2vB_ |
|
not modelled |
92.9 |
21 |
PDB header:electron transport
Chain: B: PDB Molecule:cytochrome f;
PDBTitle: n153q mutant of cytochrome f from chlamydomonas reinhardtii
|
|
|
| 76 | c4iqzD_ |
|
not modelled |
92.8 |
27 |
PDB header:unknown function
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: the crystal structure of a large insert in rna polymerase (rpoc)2 subunit from e. coli
|
|
|
| 77 | c3oamD_ |
|
not modelled |
92.6 |
10 |
PDB header:transferase
Chain: D: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: crystal structure of cytidylyltransferase from vibrio cholerae
|
|
|
| 78 | c4cgkA_ |
|
not modelled |
92.3 |
9 |
PDB header:cell cycle
Chain: A: PDB Molecule:secreted 45 kda protein;
PDBTitle: crystal structure of the essential protein pcsb from streptococcus2 pneumoniae
|
|
|
| 79 | c1ctmA_ |
|
not modelled |
92.3 |
19 |
PDB header:electron transport(cytochrome)
Chain: A: PDB Molecule:cytochrome f;
PDBTitle: crystal structure of chloroplast cytochrome f reveals a2 novel cytochrome fold and unexpected heme ligation
|
|
|
| 80 | d1w77a1 |
|
not modelled |
92.3 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 81 | d1brwa3 |
|
not modelled |
91.1 |
27 |
Fold:alpha/beta-Hammerhead
Superfamily:Pyrimidine nucleoside phosphorylase C-terminal domain
Family:Pyrimidine nucleoside phosphorylase C-terminal domain |
|
|
| 82 | c1q90A_ |
|
not modelled |
90.9 |
20 |
PDB header:photosynthesis
Chain: A: PDB Molecule:apocytochrome f;
PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
|
|
|
| 83 | c3d5nB_ |
|
not modelled |
90.4 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:q97w15_sulso;
PDBTitle: crystal structure of the q97w15_sulso protein from sulfolobus2 solfataricus. nesg target ssr125.
|
|
|
| 84 | d1dcza_ |
|
not modelled |
90.2 |
22 |
Fold:Barrel-sandwich hybrid
Superfamily:Single hybrid motif
Family:Biotinyl/lipoyl-carrier proteins and domains |
|
|
| 85 | c2vshB_ |
|
not modelled |
90.1 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate
PDBTitle: synthesis of cdp-activated ribitol for teichoic acid2 precursors in streptococcus pneumoniae
|
|
|
| 86 | c2jxmB_ |
|
not modelled |
89.7 |
29 |
PDB header:electron transport
Chain: B: PDB Molecule:cytochrome f;
PDBTitle: ensemble of twenty structures of the prochlorothrix2 hollandica plastocyanin- cytochrome f complex
|
|
|
| 87 | c2kccA_ |
|
not modelled |
89.4 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: solution structure of biotinoyl domain from human acetyl-2 coa carboxylase 2
|
|
|
| 88 | c2ejgD_ |
|
not modelled |
89.1 |
22 |
PDB header:ligase
Chain: D: PDB Molecule:149aa long hypothetical methylmalonyl-coa decarboxylase
PDBTitle: crystal structure of the biotin protein ligase (mutation r48a) and2 biotin carboxyl carrier protein complex from pyrococcus horikoshii3 ot3
|
|
|
| 89 | d2tpta3 |
|
not modelled |
88.4 |
25 |
Fold:alpha/beta-Hammerhead
Superfamily:Pyrimidine nucleoside phosphorylase C-terminal domain
Family:Pyrimidine nucleoside phosphorylase C-terminal domain |
|
|
| 90 | c3okrA_ |
|
not modelled |
87.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
|
|
| 91 | c1tu2B_ |
|
not modelled |
87.0 |
33 |
PDB header:electron transport
Chain: B: PDB Molecule:apocytochrome f;
PDBTitle: the complex of nostoc cytochrome f and plastocyanin determin with2 paramagnetic nmr. based on the structures of cytochrome f and3 plastocyanin, 10 structures
|
|
|
| 92 | c6ifdD_ |
|
not modelled |
86.6 |
10 |
PDB header:sugar binding protein
Chain: D: PDB Molecule:cmp-n-acetylneuraminate synthetase;
PDBTitle: crystal structure of cmp-n-acetylneuraminate synthetase from vibrio2 cholerae in complex with cdp and mg2+.
|
|
|
| 93 | d1uoua3 |
|
not modelled |
86.1 |
20 |
Fold:alpha/beta-Hammerhead
Superfamily:Pyrimidine nucleoside phosphorylase C-terminal domain
Family:Pyrimidine nucleoside phosphorylase C-terminal domain |
|
|
| 94 | c4xwiA_ |
|
not modelled |
85.0 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: x-ray crystal structure of cmp-kdo synthase from pseudomonas2 aeruginosa
|
|
|
| 95 | c5c22A_ |
|
not modelled |
84.8 |
17 |
PDB header:protein transport
Chain: A: PDB Molecule:chromosomal hemolysin d;
PDBTitle: crystal structure of zn-bound hlyd from e. coli
|
|
|
| 96 | c3tw6B_ |
|
not modelled |
84.7 |
19 |
PDB header:ligase/activator
Chain: B: PDB Molecule:pyruvate carboxylase protein;
PDBTitle: structure of rhizobium etli pyruvate carboxylase t882a with the2 allosteric activator, acetyl coenzyme-a
|
|
|
| 97 | c4y7uA_ |
|
not modelled |
84.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotidyl transferase;
PDBTitle: structural analysis of muru
|
|
|
| 98 | c5xu0B_ |
|
not modelled |
84.4 |
26 |
PDB header:transport protein
Chain: B: PDB Molecule:membrane-fusion protein;
PDBTitle: structure of the membrane fusion protein spr0693 from streptococcus2 pneumoniae r6
|
|
|
| 99 | c3polA_ |
|
not modelled |
84.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: 2.3 angstrom crystal structure of 3-deoxy-manno-octulosonate2 cytidylyltransferase (kdsb) from acinetobacter baumannii.
|
|
|
| 100 | d1tu2b2 |
|
not modelled |
83.7 |
31 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |
|
|
| 101 | c3f1cB_ |
|
not modelled |
83.4 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:putative 2-c-methyl-d-erythritol 4-phosphate
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from listeria monocytogenes
|
|
|
| 102 | c2j0fC_ |
|
not modelled |
83.2 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
|
|
|
| 103 | c1otpA_ |
|
not modelled |
83.1 |
25 |
PDB header:phosphorylase
Chain: A: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
|
|
|
| 104 | c4p6vA_ |
|
not modelled |
82.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:na(+)-translocating nadh-quinone reductase subunit a;
PDBTitle: crystal structure of the na+-translocating nadh: ubiquinone2 oxidoreductase from vibrio cholerae
|
|
|
| 105 | c4mybA_ |
|
not modelled |
82.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of francisella tularensis 2-c-methyl-d-erythritol 4-2 phosphate cytidylyltransferase (ispd)
|
|
|
| 106 | c2b8gA_ |
|
not modelled |
82.4 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:biotin/lipoyl attachment protein;
PDBTitle: solution structure of bacillus subtilis blap biotinylated-2 form (energy minimized mean structure)
|
|
|
| 107 | c3h5qA_ |
|
not modelled |
81.8 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside phosphorylase;
PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
|
|
|
| 108 | c2aujD_ |
|
not modelled |
80.7 |
29 |
PDB header:transferase
Chain: D: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
|
|
|
| 109 | c4jisB_ |
|
not modelled |
80.3 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:ribitol-5-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ribitol 5-phosphate cytidylyltransferase (tari)2 from bacillus subtilis
|
|
|
| 110 | c5xhwA_ |
|
not modelled |
80.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:putative 6-deoxy-d-mannoheptose pathway protein;
PDBTitle: crystal structure of hddc from yersinia pseudotuberculosis
|
|
|
| 111 | c4l8jA_ |
|
not modelled |
79.8 |
22 |
PDB header:transport protein
Chain: A: PDB Molecule:putative efflux transporter;
PDBTitle: crystal structure of a putative efflux transporter (bacegg_01895) from2 bacteroides eggerthii dsm 20697 at 2.06 a resolution
|
|
|
| 112 | c3bg3A_ |
|
not modelled |
79.7 |
28 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
| 113 | c1t5eB_ |
|
not modelled |
79.6 |
26 |
PDB header:transport protein
Chain: B: PDB Molecule:multidrug resistance protein mexa;
PDBTitle: the structure of mexa
|
|
|
| 114 | c4tkoB_ |
|
not modelled |
79.5 |
26 |
PDB header:membrane protein
Chain: B: PDB Molecule:emra;
PDBTitle: structure of the periplasmic adaptor protein emra
|
|
|
| 115 | d1fxoa_ |
|
not modelled |
79.2 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 116 | d1bdoa_ |
|
not modelled |
78.8 |
16 |
Fold:Barrel-sandwich hybrid
Superfamily:Single hybrid motif
Family:Biotinyl/lipoyl-carrier proteins and domains |
|
|
| 117 | c2k33A_ |
|
not modelled |
78.4 |
22 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:acra;
PDBTitle: solution structure of an n-glycosylated protein using in2 vitro glycosylation
|
|
|
| 118 | d1qwja_ |
|
not modelled |
78.2 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 119 | d1o78a_ |
|
not modelled |
77.9 |
17 |
Fold:Barrel-sandwich hybrid
Superfamily:Single hybrid motif
Family:Biotinyl/lipoyl-carrier proteins and domains |
|
|
| 120 | c3lnnB_ |
|
not modelled |
77.8 |
35 |
PDB header:metal transport
Chain: B: PDB Molecule:membrane fusion protein (mfp) heavy metal cation efflux
PDBTitle: crystal structure of zneb from cupriavidus metallidurans
|
|
|