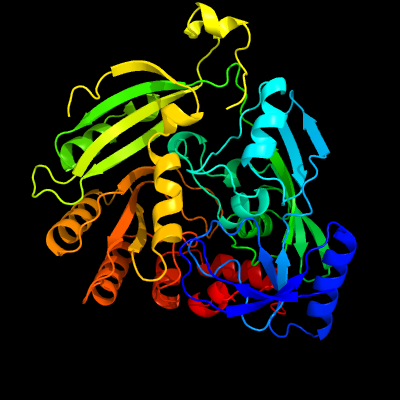
| 1 | c4fdoA_
|
|
 |
100.0 |
100 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:oxidoreductase dpre1;
PDBTitle: mycobacterium tuberculosis dpre1 in complex with ct319
|
|
|
|
| 2 | c2vfvA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xylitol oxidase;
PDBTitle: alditol oxidase from streptomyces coelicolor a3(2): complex2 with sulphite
|
|
|
|
| 3 | c6c80B_
|
|
 |
100.0 |
18 |
PDB header:immune system
Chain: B: PDB Molecule:cytokinin oxidase luckx1.1;
PDBTitle: crystal structure of a flax cytokinin oxidase
|
|
|
|
| 4 | c4ml8C_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytokinin oxidase 2;
PDBTitle: structure of maize cytokinin oxidase/dehydrogenase 2 (zmcko2)
|
|
|
|
| 5 | c3bw7A_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 1;
PDBTitle: maize cytokinin oxidase/dehydrogenase complexed with the allenic2 cytokinin analog ha-1
|
|
|
|
| 6 | c3pm9A_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution
|
|
|
|
| 7 | c4oalB_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytokinin dehydrogenase 4;
PDBTitle: crystal structure of maize cytokinin oxidase/dehydrogenase 4 (zmcko4)2 in complex with phenylurea inhibitor cppu in alternative spacegroup
|
|
|
|
| 8 | c2bvfA_
|
|
 |
100.0 |
18 |
PDB header:oxidase
Chain: A: PDB Molecule:6-hydroxy-d-nicotine oxidase;
PDBTitle: crystal structure of 6-hydoxy-d-nicotine oxidase from2 arthrobacter nicotinovorans. crystal form 3 (p1)
|
|
|
|
| 9 | c1i19B_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase from b.sterolicum
|
|
|
|
| 10 | c3vteA_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tetrahydrocannabinolic acid synthase;
PDBTitle: crystal structure of tetrahydrocannabinolic acid synthase from2 cannabis sativa
|
|
|
|
| 11 | c4bc9C_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase, peroxisomal;
PDBTitle: mammalian alkyldihydroxyacetonephosphate synthase: wild-type, adduct2 with cyanoethyl
|
|
|
|
| 12 | c2exrA_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 7;
PDBTitle: x-ray structure of cytokinin oxidase/dehydrogenase (ckx) from2 arabidopsis thaliana at5g21482
|
|
|
|
| 13 | c3w8wA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative fad-dependent oxygenase encm;
PDBTitle: the crystal structure of encm
|
|
|
|
| 14 | c3js8A_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: solvent-stable cholesterol oxidase
|
|
|
|
| 15 | c1wveB_
|
|
 |
100.0 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-cresol dehydrogenase [hydroxylating]
PDBTitle: p-cresol methylhydroxylase: alteration of the structure of2 the flavoprotein subunit upon its binding to the3 cytochrome subunit
|
|
|
|
| 16 | c5l6fA_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad linked oxidase-like protein;
PDBTitle: xylooligosaccharide oxidase from myceliophthora thermophila c1 in2 complex with xylobiose
|
|
|
|
| 17 | c4ud8B_
|
|
 |
100.0 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fad-binding and bbe domain-containing protein;
PDBTitle: atbbe15
|
|
|
|
| 18 | c6eo5A_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ppbbe-like 1 d396n;
PDBTitle: physcomitrella patens bbe-like 1 variant d396n
|
|
|
|
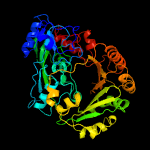
| 19 | c3tsjA_
|
|
 |
100.0 |
13 |
PDB header:allergen, oxidoreductase
Chain: A: PDB Molecule:pollen allergen phl p 4;
PDBTitle: crystal structure of phl p 4, a grass pollen allergen with glucose2 dehydrogenase activity
|
|
|
|
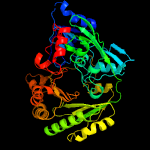
| 20 | c3rjaA_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbohydrate oxidase;
PDBTitle: crystal structure of carbohydrate oxidase from microdochium nivale in2 complex with substrate analogue
|
|
|
|
| 21 | c1zr6A_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucooligosaccharide oxidase;
PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
|
|
|
| 22 | c6f73B_ |
|
not modelled |
100.0 |
15 |
PDB header:flavoprotein
Chain: B: PDB Molecule:mtvao615;
PDBTitle: crystal structure of vao-type flavoprotein mtvao615 at ph 5.0 from2 myceliophthora thermophila c1
|
|
|
| 23 | c6f74B_ |
|
not modelled |
100.0 |
14 |
PDB header:flavoprotein
Chain: B: PDB Molecule:alcohol oxidase;
PDBTitle: crystal structure of vao-type flavoprotein mtvao713 from2 myceliophthora thermophila c1
|
|
|
| 24 | c3fwaA_ |
|
not modelled |
100.0 |
15 |
PDB header:flavoprotein
Chain: A: PDB Molecule:reticuline oxidase;
PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
|
|
|
| 25 | c3d2hA_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:berberine bridge-forming enzyme;
PDBTitle: structure of berberine bridge enzyme from eschscholzia californica,2 monoclinic crystal form
|
|
|
| 26 | c5d79B_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:berberine bridge enzyme-like protein;
PDBTitle: structure of bbe-like #28 from arabidopsis thaliana
|
|
|
| 27 | c3popD_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gilr oxidase;
PDBTitle: the crystal structure of gilr, an oxidoreductase that catalyzes the2 terminal step of gilvocarcin biosynthesis
|
|
|
| 28 | c5i1wD_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:crmk;
PDBTitle: crystal structure of crmk, a flavoenzyme involved in the shunt product2 recycling mechanism in caerulomycin biosynthesis
|
|
|
| 29 | c5fxpA_ |
|
not modelled |
100.0 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol oxidase;
PDBTitle: crystal structure of eugenol oxidase in complex with2 vanillin
|
|
|
| 30 | c2y3rC_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:taml;
PDBTitle: structure of the tirandamycin-bound fad-dependent tirandamycin oxidase2 taml in p21 space group
|
|
|
| 31 | c2ipiD_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aclacinomycin oxidoreductase (aknox);
PDBTitle: crystal structure of aclacinomycin oxidoreductase
|
|
|
| 32 | c2wdwB_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative hexose oxidase;
PDBTitle: the native crystal structure of the primary hexose oxidase (2 dbv29) in antibiotic a40926 biosynthesis
|
|
|
| 33 | c1ahuB_ |
|
not modelled |
100.0 |
11 |
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
|
|
|
| 34 | c2uuvC_ |
|
not modelled |
100.0 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase;
PDBTitle: alkyldihydroxyacetonephosphate synthase in p1
|
|
|
| 35 | c1f0xA_ |
|
not modelled |
100.0 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral membrane2 respiratory enzyme.
|
|
|
| 36 | d1w1oa2 |
|
not modelled |
100.0 |
27 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
|
|
| 37 | d2i0ka2 |
|
not modelled |
100.0 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
|
|
| 38 | d1wvfa2 |
|
not modelled |
100.0 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
|
|
| 39 | d1e8ga2 |
|
not modelled |
100.0 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
|
|
| 40 | d1f0xa2 |
|
not modelled |
100.0 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
|
|
| 41 | d1uxya1 |
|
not modelled |
100.0 |
16 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
|
|
| 42 | d1hska1 |
|
not modelled |
100.0 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
|
|
| 43 | c4pytA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of a murb family ep-udp-n-acetylglucosamine2 reductase
|
|
|
| 44 | c4jayC_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of p. aeruginosa murb in complex with nadp+
|
|
|
| 45 | c1hskA_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of s. aureus murb
|
|
|
| 46 | c1mbbA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uridine diphospho-n-acetylenolpyruvylglucosamine
PDBTitle: oxidoreductase
|
|
|
| 47 | c2yvsA_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycolate oxidase subunit glce;
PDBTitle: crystal structure of glycolate oxidase subunit glce from thermus2 thermophilus hb8
|
|
|
| 48 | c3i99A_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: the crystal structure of the udp-n-acetylenolpyruvoylglucosamine2 reductase from the vibrio cholerae o1 biovar tor
|
|
|
| 49 | c5jzxB_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of udp-n-acetylenolpyruvoylglucosamine reductase2 (murb) from mycobacterium tuberculosis
|
|
|
| 50 | c2gquA_ |
|
not modelled |
99.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvylglucosamine reductase;
PDBTitle: crystal structure of udp-n-acetylenolpyruvylglucosamine2 reductase (murb) from thermus caldophilus
|
|
|
| 51 | d2i0ka1 |
|
not modelled |
99.6 |
10 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cholesterol oxidase |
|
|
| 52 | d1w1oa1 |
|
not modelled |
98.5 |
10 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cytokinin dehydrogenase 1 |
|
|
| 53 | d1e8ga1 |
|
not modelled |
98.3 |
10 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
|
|
| 54 | d1wvfa1 |
|
not modelled |
98.2 |
10 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
|
|
| 55 | d1ffvc2 |
|
not modelled |
97.9 |
16 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 56 | c5y6qB_ |
|
not modelled |
97.4 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aldehyde oxidase medium subunit;
PDBTitle: crystal structure of an aldehyde oxidase from methylobacillus sp.2 ky4400
|
|
|
| 57 | c1t3qF_ |
|
not modelled |
97.4 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:quinoline 2-oxidoreductase medium subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
|
|
| 58 | c1ffuF_ |
|
not modelled |
97.4 |
17 |
PDB header:hydrolase
Chain: F: PDB Molecule:cutm, flavoprotein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
|
|
| 59 | c1n62C_ |
|
not modelled |
97.4 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbon monoxide dehydrogenase medium chain;
PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
|
|
|
| 60 | d1v97a6 |
|
not modelled |
97.3 |
10 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 61 | c3hrdC_ |
|
not modelled |
97.3 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinate dehydrogenase fad-subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
|
|
| 62 | d1t3qc2 |
|
not modelled |
97.3 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 63 | d3b9jb2 |
|
not modelled |
97.2 |
13 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 64 | d1n62c2 |
|
not modelled |
97.1 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 65 | c4zohB_ |
|
not modelled |
97.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase fad-binding subunit;
PDBTitle: crystal structure of glyceraldehyde oxidoreductase
|
|
|
| 66 | c5g5hB_ |
|
not modelled |
96.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative xanthine dehydrogenase yags fad-binding subunit;
PDBTitle: escherichia coli periplasmic aldehyde oxidase r440h mutant
|
|
|
| 67 | c3etrM_ |
|
not modelled |
96.8 |
8 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of xanthine oxidase in complex with lumazine
|
|
|
| 68 | c3b9jJ_ |
|
not modelled |
96.7 |
8 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:xanthine oxidase;
PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
|
|
|
| 69 | d1f0xa1 |
|
not modelled |
96.7 |
8 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:D-lactate dehydrogenase |
|
|
| 70 | d1jroa4 |
|
not modelled |
96.6 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 71 | c1rm6E_ |
|
not modelled |
96.4 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:4-hydroxybenzoyl-coa reductase beta subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
|
|
| 72 | c2w3rG_ |
|
not modelled |
96.2 |
18 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
|
|
|
| 73 | c1wygA_ |
|
not modelled |
96.0 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
|
|
|
| 74 | d1rm6b2 |
|
not modelled |
95.8 |
15 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
|
|
| 75 | c4uhxA_ |
|
not modelled |
93.4 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidase;
PDBTitle: human aldehyde oxidase in complex with phthalazine and thioridazine
|
|
|
| 76 | c3zyvA_ |
|
not modelled |
92.8 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aox3;
PDBTitle: crystal structure of the mouse liver aldehyde oxidase 3 (maox3)
|
|
|
| 77 | c2uvaI_ |
|
not modelled |
91.0 |
23 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
|
|
| 78 | c2vkzH_ |
|
not modelled |
89.1 |
20 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid synthase type i2 multienzyme complex
|
|
|
| 79 | c4b3yB_ |
|
not modelled |
79.1 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: cryo-em structure of the mycobacterial fatty acid synthase
|
|
|
| 80 | c2gvsA_ |
|
not modelled |
72.7 |
24 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:chemosensory protein csp-sg4;
PDBTitle: nmr solution structure of cspsg4
|
|
|
| 81 | d1n8va_ |
|
not modelled |
69.6 |
21 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
|
|
| 82 | d1kx9b_ |
|
not modelled |
69.3 |
21 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
|
|
| 83 | c1jk9D_ |
|
not modelled |
61.0 |
7 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:copper chaperone for superoxide dismutase;
PDBTitle: heterodimer between h48f-ysod1 and yccs
|
|
|
| 84 | c1qupA_ |
|
not modelled |
60.3 |
7 |
PDB header:chaperone
Chain: A: PDB Molecule:superoxide dismutase 1 copper chaperone;
PDBTitle: crystal structure of the copper chaperone for superoxide dismutase
|
|
|
| 85 | d1itza2 |
|
not modelled |
55.6 |
12 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like Pyr module |
|
|
| 86 | c2bp7F_ |
|
not modelled |
41.1 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:2-oxoisovalerate dehydrogenase beta subunit;
PDBTitle: new crystal form of the pseudomonas putida branched-chain2 dehydrogenase (e1)
|
|
|
| 87 | c2r8pA_ |
|
not modelled |
40.1 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:transketolase 1;
PDBTitle: transketolase from e. coli in complex with substrate d-fructose-6-2 phosphate
|
|
|
| 88 | d2r8oa1 |
|
not modelled |
39.8 |
4 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like Pyr module |
|
|
| 89 | d1umdb1 |
|
not modelled |
38.9 |
7 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
|
|
| 90 | c4wcxC_ |
|
not modelled |
37.3 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:biotin and thiamin synthesis associated;
PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
|
|
|
| 91 | c4usiC_ |
|
not modelled |
35.5 |
14 |
PDB header:signaling protein
Chain: C: PDB Molecule:nitrogen regulatory protein pii;
PDBTitle: nitrogen regulatory protein pii from chlamydomonas2 reinhardtii in complex with mgatp and 2-oxoglutarate
|
|
|
| 92 | c3komB_ |
|
not modelled |
35.2 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:transketolase;
PDBTitle: crystal structure of apo transketolase from francisella tularensis
|
|
|
| 93 | c6je8A_ |
|
not modelled |
34.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of a beta-n-acetylhexosaminidase
|
|
|
| 94 | c4r25A_ |
|
not modelled |
34.6 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:nitrogen regulatory pii-like protein;
PDBTitle: structure of b. subtilis glnk
|
|
|
| 95 | d1qupa2 |
|
not modelled |
34.6 |
9 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
|
|
| 96 | c5oarB_ |
|
not modelled |
33.3 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of native beta-n-acetylhexosaminidase isolated from2 aspergillus oryzae
|
|
|
| 97 | c3ncpD_ |
|
not modelled |
33.0 |
18 |
PDB header:signaling protein
Chain: D: PDB Molecule:nitrogen regulatory protein p-ii (glnb-2);
PDBTitle: glnk2 from archaeoglobus fulgidus
|
|
|
| 98 | c3bzqA_ |
|
not modelled |
32.8 |
24 |
PDB header:signaling protein/transcription
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii;
PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
|
|
|
| 99 | d1jaka1 |
|
not modelled |
32.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
|
|
| 100 | c3hylB_ |
|
not modelled |
32.5 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:transketolase;
PDBTitle: crystal structure of transketolase from bacillus anthracis
|
|
|
| 101 | c2rd5D_ |
|
not modelled |
31.9 |
18 |
PDB header:protein binding
Chain: D: PDB Molecule:pii protein;
PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
|
|
|
| 102 | c2crlA_ |
|
not modelled |
31.8 |
5 |
PDB header:chaperone
Chain: A: PDB Molecule:copper chaperone for superoxide dismutase;
PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
|
|
|
| 103 | d1gpua2 |
|
not modelled |
31.3 |
14 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like Pyr module |
|
|
| 104 | d1w85b1 |
|
not modelled |
30.9 |
15 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
|
|
| 105 | c4h04B_ |
|
not modelled |
30.3 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:lacto-n-biosidase;
PDBTitle: lacto-n-biosidase from bifidobacterium bifidum
|
|
|
| 106 | d1ik6a1 |
|
not modelled |
29.9 |
15 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |
|
|
| 107 | d2bi7a1 |
|
not modelled |
29.4 |
16 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:UDP-galactopyranose mutase, N-terminal domain |
|
|
| 108 | c2yl8A_ |
|
not modelled |
29.4 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
|
|
| 109 | c4c7vA_ |
|
not modelled |
29.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:transketolase;
PDBTitle: apo transketolase from lactobacillus salivarius at 2.2a resolution
|
|
|
| 110 | d2piia_ |
|
not modelled |
29.2 |
17 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
|
|
| 111 | c6a8mA_ |
|
not modelled |
29.0 |
10 |
PDB header:dna binding protein
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: n-terminal domain of fact complex subunit spt16 from eremothecium2 gossypii (ashbya gossypii)
|
|
|
| 112 | c4ozlA_ |
|
not modelled |
28.7 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:nitrogen regulatory protein p-ii;
PDBTitle: glnk2 from haloferax mediterranei complexed with amp
|
|
|
| 113 | c3nsnA_ |
|
not modelled |
28.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylglucosaminidase;
PDBTitle: crystal structure of insect beta-n-acetyl-d-hexosaminidase ofhex12 complexed with tmg-chitotriomycin
|
|
|
| 114 | c3m7iA_ |
|
not modelled |
28.2 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:transketolase;
PDBTitle: crystal structure of transketolase in complex with thiamine2 diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion
|
|
|
| 115 | d1yhta1 |
|
not modelled |
27.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-N-acetylhexosaminidase catalytic domain |
|
|
| 116 | c2ylaA_ |
|
not modelled |
26.7 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
|
|
|
| 117 | d1o6da_ |
|
not modelled |
26.3 |
11 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:YbeA-like |
|
|
| 118 | d1poib_ |
|
not modelled |
26.3 |
26 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase beta subunit-like |
|
|
| 119 | d1fe0a_ |
|
not modelled |
25.9 |
28 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
|
|
| 120 | c3gh7A_ |
|
not modelled |
25.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of beta-hexosaminidase from paenibacillus sp. ts122 in complex with galnac
|
|
|