1 c5f15A_
94.0
13
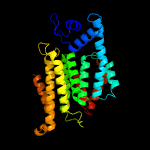
PDB header: transferaseChain: A: PDB Molecule: 4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
2 c6p25A_
73.7
13
PDB header: transferaseChain: A: PDB Molecule: dolichyl-phosphate-mannose--protein mannosyltransferase 1;PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
3 c3wajA_
72.9
13
PDB header: transferaseChain: A: PDB Molecule: transmembrane oligosaccharyl transferase;PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
4 c4ymkA_
69.4
12
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa desaturase 1;PDBTitle: crystal structure of stearoyl-coenzyme a desaturase 1
5 c6eznF_
57.7
12
PDB header: membrane proteinChain: F: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
6 c4fypA_
57.0
17
PDB header: plant proteinChain: A: PDB Molecule: vegetative storage protein 1;PDBTitle: crystal structure of plant vegetative storage protein
7 c1okgA_
53.9
13
PDB header: transferaseChain: A: PDB Molecule: possible 3-mercaptopyruvate sulfurtransferase;PDBTitle: 3-mercaptopyruvate sulfurtransferase from leishmania major
8 c4dh4A_
50.6
23
PDB header: isomeraseChain: A: PDB Molecule: mif;PDBTitle: macrophage migration inhibitory factor toxoplasma gondii
9 c5ir6B_
50.4
11
PDB header: oxidoreductaseChain: B: PDB Molecule: bd-type quinol oxidase subunit ii;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
10 c4nsmA_
48.1
43
PDB header: structural proteinChain: A: PDB Molecule: collagen-like protein sclb;PDBTitle: crystal structure of the streptococcal collagen-like protein 22 globular domain from invasive m3-type group a streptococcus
11 c3utnX_
45.9
22
PDB header: transferaseChain: X: PDB Molecule: thiosulfate sulfurtransferase tum1;PDBTitle: crystal structure of tum1 protein from saccharomyces cerevisiae
12 c6cuqB_
45.9
11
PDB header: cytokineChain: B: PDB Molecule: macrophage migration inhibitory factor-like protein;PDBTitle: crystal structure of macrophage migration inhibitory factor-like2 protein (ehmif) from entamoeba histolytica
13 d1y5ic1
42.1
24
Fold: Heme-binding four-helical bundleSuperfamily: Respiratory nitrate reductase 1 gamma chainFamily: Respiratory nitrate reductase 1 gamma chain
14 c2m7gA_
38.6
24
PDB header: cell adhesion, structural protein, electChain: A: PDB Molecule: geopilin domain 1 protein;PDBTitle: structure of the type iva major pilin from the electrically conductive2 bacterial nanowires of geobacter sulfurreducens
15 d2ji7a3
38.1
15
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase PP module
16 d1ypze2
38.0
37
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)
17 c5un4C_
37.8
17
PDB header: hydrolaseChain: C: PDB Molecule: 4-oxalocrotonate tautomerase;PDBTitle: crystal structure of native fused 4-ot
18 c1yo8A_
37.7
26
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-2;PDBTitle: structure of the c-terminal domain of human thrombospondin-2
19 c1ux6A_
36.2
30
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-1;PDBTitle: structure of a thrombospondin c-terminal fragment reveals a novel2 calcium core in the type 3 repeats
20 c3zm8A_
36.0
43
PDB header: hydrolaseChain: A: PDB Molecule: gh26 endo-beta-1,4-mannanase;PDBTitle: crystal structure of podospora anserina gh26-cbm352 beta-(1,4)-mannanase
21 c3sy6A_
not modelled
34.4
37
PDB header: cell adhesionChain: A: PDB Molecule: fimbrial protein bf1861;PDBTitle: crystal structure of a fimbrial protein bf1861 [bacteroides fragilis2 nctc 9343] (bf1861) from bacteroides fragilis nctc 9343 at 1.90 a3 resolution
22 d2gdga1
not modelled
33.8
20
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related
23 c3fbyC_
not modelled
33.7
34
PDB header: cell adhesionChain: C: PDB Molecule: cartilage oligomeric matrix protein;PDBTitle: the crystal structure of the signature domain of cartilage oligomeric2 matrix protein.
24 d2d9ra1
not modelled
32.9
30
Fold: Double-split beta-barrelSuperfamily: AF2212/PG0164-likeFamily: PG0164-like
25 d1gd0a_
not modelled
32.1
18
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related
26 c4b8yA_
not modelled
32.0
11
PDB header: transport protein/siderophoreChain: A: PDB Molecule: fhud2;PDBTitle: ferrichrome-bound fhud2
27 c4p55A_
not modelled
31.6
29
PDB header: dna binding proteinChain: A: PDB Molecule: viral irf2-like protein;PDBTitle: crystal structure of dna binding domain of k11 from kshv
28 d1hxma2
not modelled
30.8
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)
29 c6ntwA_
not modelled
30.7
17
PDB header: transferase/transferase inhibitorChain: A: PDB Molecule: probable l,d-transpeptidase ycbb;PDBTitle: crystal structure of e. coli ycbb
30 c3rceA_
not modelled
30.6
12
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
31 c3ufiA_
not modelled
30.5
33
PDB header: cell adhesionChain: A: PDB Molecule: hypothetical protein bacova_04980;PDBTitle: crystal structure of a putative cell adhesion protein (bacova_04980)2 from bacteroides ovatus atcc 8483 at 2.18 a resolution
32 c4v3aC_
not modelled
29.8
75
PDB header: transport proteinChain: C: PDB Molecule: pleurotolysin b;PDBTitle: membrane bound pleurotolysin prepore (tmh1 lock) trapped with2 engineered disulphide cross-link
33 c6hpfA_
not modelled
29.6
60
PDB header: hydrolaseChain: A: PDB Molecule: endo-b-mannanase;PDBTitle: structure of inactive e165q mutant of fungal non-cbm carrying gh262 endo-b-mannanase from yunnania penicillata in complex with alpha-62-3 61-di-galactosyl-mannotriose
34 c6basA_
not modelled
28.3
26
PDB header: transferaseChain: A: PDB Molecule: peptidoglycan glycosyltransferase roda;PDBTitle: crystal structure of thermus thermophilus rod shape determining2 protein roda d255a mutant (q5six3_thet8)
35 c4m1eC_
not modelled
27.7
12
PDB header: transferaseChain: C: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: crystal structure of purine nucleoside phosphorylase i from2 planctomyces limnophilus dsm 3776, nysgrc target 029364.
36 c3kopB_
not modelled
27.7
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
37 c3tijA_
not modelled
27.3
16
PDB header: membrane proteinChain: A: PDB Molecule: nupc family protein;PDBTitle: crystal structure of a concentrative nucleoside transporter from2 vibrio cholerae
38 c3fwtA_
not modelled
27.2
26
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: crystal structure of leishmania major mif2
39 c2oq2B_
not modelled
27.2
21
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphoadenosine phosphosulfate reductase;PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
40 c5flyB_
not modelled
27.0
13
PDB header: metal transportChain: B: PDB Molecule: ferrichrome-binding periplasmic protein;PDBTitle: the fhud protein from s.pseudintermedius
41 c4k4kA_
not modelled
26.6
42
PDB header: cell adhesionChain: A: PDB Molecule: putative cell adhesion protein;PDBTitle: crystal structure of a putative cell adhesion protein (bacuni_00621)2 from bacteroides uniformis atcc 8492 at 1.67 a resolution
42 c3caiA_
not modelled
26.0
18
PDB header: transferaseChain: A: PDB Molecule: possible aminotransferase;PDBTitle: crystal structure of mycobacterium tuberculosis rv3778c2 protein
43 c5l2bC_
not modelled
25.7
19
PDB header: transport proteinChain: C: PDB Molecule: nucleoside permease;PDBTitle: structure of cntnw n149s, e332a in an outward-facing state
44 d1ulza2
not modelled
25.7
17
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
45 d1ku0a_
not modelled
25.6
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase
46 c5zbhA_
not modelled
25.5
15
PDB header: signaling proteinChain: A: PDB Molecule: neuropeptide y receptor type 1,t4 lysozyme,neuropeptide yPDBTitle: the crystal structure of human neuropeptide y y1 receptor with bms-2 193885
47 c6c14A_
not modelled
25.5
19
PDB header: membrane protein, metal transportChain: A: PDB Molecule: protocadherin-15;PDBTitle: cryoem structure of mouse pcdh15-1ec-lhfpl5 complex
48 c4tvrA_
not modelled
25.4
17
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase uhrf2;PDBTitle: tandem tudor and phd domains of uhrf2
49 c5wt2A_
not modelled
24.8
11
PDB header: transferaseChain: A: PDB Molecule: cysteine desulfurase iscs;PDBTitle: nifs from helicobacter pylori
50 d1n2za_
not modelled
24.1
9
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
51 c4or2A_
not modelled
24.1
17
PDB header: signaling proteinChain: A: PDB Molecule: soluble cytochrome b562, metabotropic glutamate receptor 1;PDBTitle: human class c g protein-coupled metabotropic glutamate receptor 1 in2 complex with a negative allosteric modulator
52 d1wi0a_
not modelled
24.0
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain
53 c6nqiB_
not modelled
23.4
17
PDB header: splicingChain: B: PDB Molecule: pre-mrna splicing factor prp8;PDBTitle: prp8 rh domain from c. merolae
54 c5ip0C_
not modelled
23.4
15
PDB header: structural proteinChain: C: PDB Molecule: pha granule-associated protein;PDBTitle: pha binding protein phap (phasin)
55 c2i34B_
not modelled
23.2
28
PDB header: hydrolaseChain: B: PDB Molecule: acid phosphatase;PDBTitle: the crystal structure of class c acid phosphatase from bacillus2 anthracis with tungstate bound
56 c4ic7B_
not modelled
23.2
27
PDB header: transferaseChain: B: PDB Molecule: dual specificity mitogen-activated protein kinase kinase 5;PDBTitle: crystal structure of the erk5 kinase domain in complex with an mkk52 binding fragment
57 c2hihB_
not modelled
23.1
25
PDB header: hydrolaseChain: B: PDB Molecule: lipase 46 kda form;PDBTitle: crystal structure of staphylococcus hyicus lipase
58 c5ah1A_
not modelled
22.8
25
PDB header: hydrolaseChain: A: PDB Molecule: triacylglycerol lipase;PDBTitle: structure of esta from clostridium botulinum
59 c2bvtB_
not modelled
22.3
44
PDB header: hydrolaseChain: B: PDB Molecule: beta-1,4-mannanase;PDBTitle: the structure of a modular endo-beta-1,4-mannanase from cellulomonas2 fimi explains the product specificity of glycoside hydrolase family3 26 mannanases.
60 c6eazA_
not modelled
22.3
23
PDB header: metal binding proteinChain: A: PDB Molecule: calcium uptake protein 2, mitochondrial;PDBTitle: apo structure of the mitochondrial calcium uniporter protein micu2
61 c3siiA_
not modelled
22.0
23
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: poly(adp-ribose) glycohydrolase;PDBTitle: the x-ray crystal structure of poly(adp-ribose) glycohydrolase bound2 to the inhibitor adp-hpd from thermomonospora curvata
62 c4epzA_
not modelled
22.0
25
PDB header: transcriptionChain: A: PDB Molecule: transcription anti-terminator antagonist upxz;PDBTitle: crystal structure of a transcription anti-terminator antagonist upxz2 (bacuni_04315) from bacteroides uniformis atcc 8492 at 1.68 a3 resolution
63 d2abwa1
not modelled
21.8
12
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)
64 c3b64A_
not modelled
21.7
30
PDB header: cytokineChain: A: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
65 c3pntA_
not modelled
21.7
46
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: nad+-glycohydrolase;PDBTitle: crystal structure of the streptococcus pyogenes nad+ glycohydrolase2 spn in complex with ifs, the immunity factor for spn
66 c5ah0B_
not modelled
21.4
8
PDB header: hydrolaseChain: B: PDB Molecule: lipase;PDBTitle: structure of lipase 1 from pelosinus fermentans
67 d1ur4a_
not modelled
21.4
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
68 c3e77A_
not modelled
21.3
15
PDB header: transferaseChain: A: PDB Molecule: phosphoserine aminotransferase;PDBTitle: human phosphoserine aminotransferase in complex with plp
69 d1ji3a_
not modelled
21.2
17
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase
70 c3wdrA_
not modelled
21.2
50
PDB header: hydrolaseChain: A: PDB Molecule: beta-mannanase;PDBTitle: crystal structure of beta-mannanase from a symbiotic protist of the2 termite reticulitermes speratus complexed with gluco-manno-3 oligosaccharide
71 c4qb7A_
not modelled
21.1
33
PDB header: cell adhesionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a fimbrial protein (bvu_2522) from bacteroides2 vulgatus atcc 8482 at 2.55 a resolution
72 c5j1sB_
not modelled
21.1
17
PDB header: hydrolaseChain: B: PDB Molecule: torsin-1a-interacting protein 2;PDBTitle: torsina-lull1 complex, h. sapiens, bound to vhh-bs2
73 c4kmhB_
not modelled
21.0
43
PDB header: protein bindingChain: B: PDB Molecule: suppressor of fused homolog;PDBTitle: crystal structure of suppressor of fused d20
74 d1jmxg_
not modelled
20.6
27
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Quinohemoprotein amine dehydrogenase C chainFamily: Quinohemoprotein amine dehydrogenase C chain
75 c3t2lA_
not modelled
20.2
42
PDB header: cell adhesionChain: A: PDB Molecule: putative cell adhesion protein;PDBTitle: crystal structure of a putative cell adhesion protein (bf1858) from2 bacteroides fragilis nctc 9343 at 2.33 a resolution
76 c4c82A_
not modelled
20.1
13
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: ispf (plasmodium falciparum) unliganded structure
77 d2cyga1
not modelled
20.1
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
78 c5iwyD_
not modelled
19.9
19
PDB header: lyaseChain: D: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: crystal structure of 2-c-methyl-d-erythritol 2,4-cyclodiphosphate2 synthase from bacillus subtitis complexed with cmp and mg2+
79 c5f5nA_
not modelled
19.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: monooxygenase;PDBTitle: the structure of monooxygenase ksta11 in complex with nad and its2 substrate
80 d1w23a_
not modelled
19.7
11
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
81 d1ux6a2
not modelled
19.6
18
Fold: TSP type-3 repeatSuperfamily: TSP type-3 repeatFamily: TSP type-3 repeat
82 c5msmD_
not modelled
19.6
29
PDB header: cell cycleChain: D: PDB Molecule: sister chromatid cohesion protein dcc1;PDBTitle: structure of the dcc1-ctf8-ctf18c trimer
83 d2fug21
not modelled
19.6
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: NQO2-like
84 c4f6oA_
not modelled
19.6
25
PDB header: hydrolaseChain: A: PDB Molecule: metacaspase-1;PDBTitle: crystal structure of the yeast metacaspase yca1
85 c6fkib_
not modelled
19.5
20
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit beta, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
86 c3cbwA_
not modelled
19.4
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ydht protein;PDBTitle: crystal structure of the ydht protein from bacillus subtilis
87 c5lgdA_
not modelled
19.4
16
PDB header: cell adhesionChain: A: PDB Molecule: platelet glycoprotein 4;PDBTitle: the cidra domain from mcvar1 pfemp1 bound to cd36
88 c6hf4A_
not modelled
19.2
50
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase family 26;PDBTitle: the structure of boman26b, a gh26 beta-mannanase from bacteroides2 ovatus, complexed with g1m4
89 c3ca8B_
not modelled
19.0
42
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein ydcf;PDBTitle: crystal structure of escherichia coli ydcf, an s-adenosyl-l-methionine2 utilizing enzyme
90 c1kyqC_
not modelled
19.0
24
PDB header: oxidoreductase, lyaseChain: C: PDB Molecule: siroheme biosynthesis protein met8;PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
91 d1okga1
not modelled
18.8
14
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)
92 d1kyqa2
not modelled
18.7
18
Fold: Siroheme synthase middle domains-likeSuperfamily: Siroheme synthase middle domains-likeFamily: Siroheme synthase middle domains-like
93 c4f7bA_
not modelled
18.4
21
PDB header: cell adhesionChain: A: PDB Molecule: lysosome membrane protein 2;PDBTitle: structure of the lysosomal domain of limp-2
94 c4r6yA_
not modelled
18.4
14
PDB header: transport proteinChain: A: PDB Molecule: putative 2-aminoethylphosphonate-binding periplasmicPDBTitle: crystal structure of solute-binding protein stm0429 from salmonella2 enterica subsp. enterica serovar typhimurium str. lt2, target efi-3 510776, a closed conformation, in complex with glycerol and acetate
95 c2wqqA_
not modelled
18.4
32
PDB header: transferaseChain: A: PDB Molecule: alpha-2,3-/2,8-sialyltransferase;PDBTitle: crystallographic analysis of monomeric cstii
96 c5v6hC_
not modelled
18.3
33
PDB header: protein bindingChain: C: PDB Molecule: pdz domain-containing protein gipc2;PDBTitle: crystal structure of myosin vi in complex with gh2 domain of gipc2
97 c4z8wA_
not modelled
18.0
23
PDB header: allergenChain: A: PDB Molecule: major pollen allergen pla l 1;PDBTitle: crystal structure of the major plantain pollen allergen pla l 1
98 c3d8kD_
not modelled
18.0
14
PDB header: hydrolaseChain: D: PDB Molecule: protein phosphatase 2c;PDBTitle: crystal structure of a phosphatase from a toxoplasma gondii
99 d1g9ka2
not modelled
18.0
26
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain