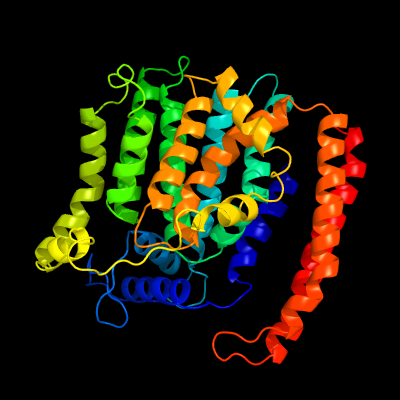
| 1 | c5f15A_
|
|
 |
99.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;
PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
|
|
|
|
| 2 | c3wajA_
|
|
 |
98.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase;
PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
|
|
|
|
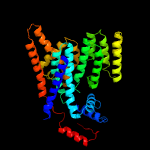
| 3 | c3rceA_
|
|
 |
98.0 |
16 |
PDB header:transferase/peptide
Chain: A: PDB Molecule:oligosaccharide transferase to n-glycosylate proteins;
PDBTitle: bacterial oligosaccharyltransferase pglb
|
|
|
|
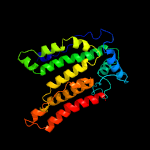
| 4 | c6p2rB_
|
|
 |
97.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 2;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
|
|
|
|
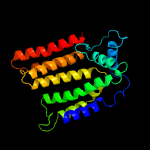
| 5 | c6p25A_
|
|
 |
97.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 1;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
|
|
|
|
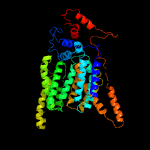
| 6 | c6eznF_
|
|
 |
96.4 |
13 |
PDB header:membrane protein
Chain: F: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
|
|
|
|
| 7 | d2d6fc2
|
|
 |
62.9 |
21 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
|
|
|
| 8 | d1zq1c2
|
|
 |
58.9 |
26 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
|
|
|
| 9 | c4j7bF_
|
|
 |
36.1 |
26 |
PDB header:transferase
Chain: F: PDB Molecule:205 kda microtubule-associated protein;
PDBTitle: crystal structure of polo-like kinase 1
|
|
|
|
| 10 | c4j7bC_
|
|
 |
36.1 |
26 |
PDB header:transferase
Chain: C: PDB Molecule:205 kda microtubule-associated protein;
PDBTitle: crystal structure of polo-like kinase 1
|
|
|
|
| 11 | c5uz9D_
|
|
 |
35.3 |
30 |
PDB header:immune system/rna
Chain: D: PDB Molecule:crispr-associated protein csy3;
PDBTitle: cryo em structure of anti-crisprs, acrf1 and acrf2, bound to type i-f2 crrna-guided crispr surveillance complex
|
|
|
|
| 12 | c4eogA_
|
|
 |
31.6 |
24 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of csx1 of pyrococcus furiosus
|
|
|
|
| 13 | d1usua_
|
|
 |
19.8 |
12 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Hsp90 middle domain |
|
|
|
| 14 | d1s4na_
|
|
 |
19.0 |
19 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Glycolipid 2-alpha-mannosyltransferase |
|
|
|
| 15 | c1s4pA_
|
|
 |
17.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:glycolipid 2-alpha-mannosyltransferase;
PDBTitle: crystal structure of yeast alpha1,2-mannosyltransferase kre2p/mnt1p:2 ternary complex with gdp/mn and methyl-alpha-mannoside acceptor
|
|
|
|
| 16 | d1g8fa2
|
|
 |
17.9 |
22 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
|
|
|
| 17 | d2prra1
|
|
 |
17.6 |
7 |
Fold:AhpD-like
Superfamily:AhpD-like
Family:Atu0492-like |
|
|
|
| 18 | c2gx8B_
|
|
 |
17.2 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:nif3-related protein;
PDBTitle: the crystal structure of bacillus cereus protein related to nif3
|
|
|
|
| 19 | c5ve9C_
|
|
 |
16.9 |
50 |
PDB header:protein binding
Chain: C: PDB Molecule:microtubule-actin cross-linking factor 1, isoforms 1/2/3/5;
PDBTitle: structure of hacf7 ef1-ef2-gar domains
|
|
|
|
| 20 | d1ht6a1
|
|
 |
16.8 |
17 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
|
|
|
| 21 | d2gx8a1 |
|
not modelled |
15.7 |
22 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
|
|
| 22 | d1v5ra1 |
|
not modelled |
15.3 |
38 |
Fold:N domain of copper amine oxidase-like
Superfamily:GAS2 domain-like
Family:GAS2 domain |
|
|
| 23 | c3arcl_ |
|
not modelled |
15.1 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
|
|
|
| 24 | c4ub6L_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
|
|
|
| 25 | c4tnhl_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
|
|
|
| 26 | c3a0bl_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of br-substituted photosystem ii complex
|
|
|
| 27 | c4il6L_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: structure of sr-substituted photosystem ii
|
|
|
| 28 | c5e7cl_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
|
|
|
| 29 | c4tnkL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
|
|
|
| 30 | c4rvyl_ |
|
not modelled |
14.6 |
33 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
|
|
|
| 31 | c4ixrl_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
|
|
|
| 32 | c4fbyL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: fs x-ray diffraction of photosystem ii
|
|
|
| 33 | c4tnil_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
|
|
|
| 34 | c4ixql_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
|
|
|
| 35 | c3wu2L_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure analysis of photosystem ii complex
|
|
|
| 36 | c4ub8l_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
|
|
|
| 37 | c4tnjL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
|
|
|
| 38 | c3arcL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
|
|
|
| 39 | c4ub6l_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
|
|
|
| 40 | c4tniL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
|
|
|
| 41 | c4ixrL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
|
|
|
| 42 | c4il6l_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: structure of sr-substituted photosystem ii
|
|
|
| 43 | c3kziL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
|
|
|
| 44 | c4ixqL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
|
|
|
| 45 | c3prrL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
|
|
|
| 46 | c4tnkl_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
|
|
|
| 47 | c5e7cL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
|
|
|
| 48 | c4ub8L_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
|
|
|
| 49 | c3a0hl_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of i-substituted photosystem ii complex
|
|
|
| 50 | c2axtL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
|
|
|
| 51 | c1s5ll_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
|
|
|
| 52 | c2axtl_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
|
|
|
| 53 | c4fbyd_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: D: PDB Molecule:photosystem ii d2 protein;
PDBTitle: fs x-ray diffraction of photosystem ii
|
|
|
| 54 | c3wu2l_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure analysis of photosystem ii complex
|
|
|
| 55 | c3bz1L_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 1 of 2). this2 file contains first monomer of psii dimer
|
|
|
| 56 | d2axtl1 |
|
not modelled |
14.6 |
33 |
Fold:Single transmembrane helix
Superfamily:Photosystem II reaction center protein L, PsbL
Family:PsbL-like |
|
|
| 57 | c1s5lL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center l protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
|
|
|
| 58 | c3prqL_ |
|
not modelled |
14.6 |
33 |
PDB header:photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
|
|
|
| 59 | c3bz2L_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 2 of 2). this2 file contains second monomer of psii dimer
|
|
|
| 60 | c4rvyL_ |
|
not modelled |
14.6 |
33 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
|
|
|
| 61 | c3a0hL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of i-substituted photosystem ii complex
|
|
|
| 62 | c4tnjl_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
|
|
|
| 63 | c4tnhL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport,photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
|
|
|
| 64 | c3a0bL_ |
|
not modelled |
14.6 |
33 |
PDB header:electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of br-substituted photosystem ii complex
|
|
|
| 65 | c4pj0l_ |
|
not modelled |
14.6 |
33 |
PDB header:oxidoreductase, electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
|
|
|
| 66 | c4pj0L_ |
|
not modelled |
14.6 |
33 |
PDB header:oxidoreductase, electron transport
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
|
|
|
| 67 | c6nd1A_ |
|
not modelled |
14.3 |
22 |
PDB header:protein transport
Chain: A: PDB Molecule:protein translocation protein sec63;
PDBTitle: cryoem structure of the sec complex from yeast
|
|
|
| 68 | d1su3a1 |
|
not modelled |
14.1 |
50 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
|
|
| 69 | c3cr8C_ |
|
not modelled |
14.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:sulfate adenylyltransferase, adenylylsulfate kinase;
PDBTitle: hexameric aps kinase from thiobacillus denitrificans
|
|
|
| 70 | d1pama3 |
|
not modelled |
13.9 |
47 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
|
|
| 71 | d1slma1 |
|
not modelled |
13.9 |
50 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
|
|
| 72 | c6hkeB_ |
|
not modelled |
13.7 |
18 |
PDB header:transport protein
Chain: B: PDB Molecule:possible tctc subunit of the tripartite tricarboxylate
PDBTitle: matc (rpa3494) from rhodopseudomonas palustris with bound malate
|
|
|
| 73 | d1t5la1 |
|
not modelled |
13.6 |
25 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
|
|
| 74 | c3jcul_ |
|
not modelled |
13.6 |
46 |
PDB header:membrane protein
Chain: L: PDB Molecule:protein photosystem ii reaction center protein l;
PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
|
|
|
| 75 | d1m55a_ |
|
not modelled |
13.4 |
15 |
Fold:Origin of replication-binding domain, RBD-like
Superfamily:Origin of replication-binding domain, RBD-like
Family:Replication protein Rep, nuclease domain |
|
|
| 76 | d1m8pa2 |
|
not modelled |
13.3 |
27 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
|
|
| 77 | c2nydB_ |
|
not modelled |
13.2 |
15 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0135 protein sa1388;
PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
|
|
|
| 78 | c3n3fB_ |
|
not modelled |
13.1 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:collagen alpha-1(xv) chain;
PDBTitle: crystal structure of the human collagen xv trimerization domain: a2 potent trimerizing unit common to multiplexin collagens
|
|
|
| 79 | d1x6va2 |
|
not modelled |
13.1 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
|
|
| 80 | c1r6xA_ |
|
not modelled |
12.7 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:atp:sulfate adenylyltransferase;
PDBTitle: the crystal structure of a truncated form of yeast atp2 sulfurylase, lacking the c-terminal aps kinase-like domain,3 in complex with sulfate
|
|
|
| 81 | d1ku1a_ |
|
not modelled |
12.7 |
12 |
Fold:alpha-alpha superhelix
Superfamily:Sec7 domain
Family:Sec7 domain |
|
|
| 82 | c3i71B_ |
|
not modelled |
12.6 |
27 |
PDB header:unknown function
Chain: B: PDB Molecule:ethanolamine utilization protein eutk;
PDBTitle: ethanolamine utilization microcompartment shell subunit, eutk c-2 terminal domain
|
|
|
| 83 | c2ka6B_ |
|
not modelled |
12.6 |
35 |
PDB header:transcription regulator
Chain: B: PDB Molecule:signal transducer and activator of transcription
PDBTitle: nmr structure of the cbp-taz2/stat1-tad complex
|
|
|
| 84 | c5a6eB_ |
|
not modelled |
12.6 |
25 |
PDB header:transport
Chain: B: PDB Molecule:pore domain of potassium channel subfamily t member 1;
PDBTitle: cryo-em structure of the slo2.2 na-activated k channel
|
|
|
| 85 | c1oftC_ |
|
not modelled |
12.4 |
14 |
PDB header:bacterial cell division inhibitor
Chain: C: PDB Molecule:hypothetical protein pa3008;
PDBTitle: crystal structure of sula from pseudomonas aeruginosa
|
|
|
| 86 | d1eaka1 |
|
not modelled |
12.3 |
50 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
|
|
| 87 | d1l6ja1 |
|
not modelled |
12.2 |
50 |
Fold:PGBD-like
Superfamily:PGBD-like
Family:MMP N-terminal domain |
|
|
| 88 | c1m8pB_ |
|
not modelled |
12.2 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:sulfate adenylyltransferase;
PDBTitle: crystal structure of p. chrysogenum atp sulfurylase in the t-state
|
|
|
| 89 | d1s0pa_ |
|
not modelled |
12.0 |
29 |
Fold:N-terminal domain of adenylylcyclase associated protein, CAP
Superfamily:N-terminal domain of adenylylcyclase associated protein, CAP
Family:N-terminal domain of adenylylcyclase associated protein, CAP |
|
|
| 90 | c6f5dI_ |
|
not modelled |
11.9 |
27 |
PDB header:hydrolase
Chain: I: PDB Molecule:atp synthase subunit epsilon, mitochondrial;
PDBTitle: trypanosoma brucei f1-atpase
|
|
|
| 91 | d1ofux_ |
|
not modelled |
11.9 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Bacterial cell division inhibitor SulA |
|
|
| 92 | c5ibwC_ |
|
not modelled |
11.7 |
13 |
PDB header:motor protein
Chain: C: PDB Molecule:myosin ic heavy chain;
PDBTitle: complex of mlcc bound to the tandem iq motif of myoc
|
|
|
| 93 | d3bmva3 |
|
not modelled |
11.6 |
47 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
|
|
| 94 | c2d6fD_ |
|
not modelled |
11.5 |
22 |
PDB header:ligase/rna
Chain: D: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit e;
PDBTitle: crystal structure of glu-trna(gln) amidotransferase in the2 complex with trna(gln)
|
|
|
| 95 | d1kona_ |
|
not modelled |
11.5 |
60 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
|
|
| 96 | c3kwpA_ |
|
not modelled |
11.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
|
|
| 97 | c1bm4A_ |
|
not modelled |
11.4 |
20 |
PDB header:viral protein
Chain: A: PDB Molecule:protein (moloney murine leukemia virus capsid);
PDBTitle: momlv capsid protein major homology region peptide analog
|
|
|
| 98 | c5a08A_ |
|
not modelled |
11.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:probable mannosyltransferase ktr4;
PDBTitle: x-ray structure of the mannosyltransferase ktr4p from s. cerevisiae
|
|
|
| 99 | d1ewna_ |
|
not modelled |
11.4 |
16 |
Fold:FMT C-terminal domain-like
Superfamily:FMT C-terminal domain-like
Family:3-methyladenine DNA glycosylase (AAG, ANPG, MPG) |
|
|