1 c4od5C_
100.0
15
PDB header: transferaseChain: C: PDB Molecule: 4-hydroxybenzoate octaprenyltransferase;PDBTitle: substrate-bound structure of a ubia homolog from aeropyrum pernix k1
2 c3dh4A_
60.9
12
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
3 c2l4fA_
32.8
42
PDB header: protein bindingChain: A: PDB Molecule: defective in cullin neddylation protein 1;PDBTitle: nmr structure of the uba domain of s. cerevisiae dcn1 bound to2 ubiquitin
4 c2xq2A_
28.1
12
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
5 d1diha2
23.9
28
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
6 c5us6L_
20.7
31
PDB header: oxidoreductaseChain: L: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: structure of dihydrodipicolinate reductase from vibrio vulnificus2 bound to nadh and 2,6 pyridine dicarboxylic acid with intact3 polyhistidine tag
7 c4o6mA_
16.9
13
PDB header: transferaseChain: A: PDB Molecule: af2299, a cdp-alcohol phosphotransferase;PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
8 c3npkB_
16.4
18
PDB header: transferaseChain: B: PDB Molecule: geranyltranstransferase;PDBTitle: the crystal structure of geranyltranstransferase from campylobacter2 jejuni
9 c4ksnC_
14.3
33
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: sdbc;PDBTitle: c-terminal domain of sdbc protein from legionella pneumophila.
10 c5tenH_
13.2
31
PDB header: oxidoreductaseChain: H: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
11 c2ww9O_
7.5
38
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l39;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the yeast2 80s ribosome
12 c3m9uD_
7.4
12
PDB header: transferaseChain: D: PDB Molecule: farnesyl-diphosphate synthase;PDBTitle: crystal structure of geranylgeranyl pyrophosphate synthase from2 lactobacillus brevis atcc 367
13 c5wwoB_
7.3
23
PDB header: rna binding proteinChain: B: PDB Molecule: essential nuclear protein 1;PDBTitle: crystal structure of enp1
14 c1drwA_
7.2
28
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
15 c6g4ww_
7.0
15
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s15a;PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state a
16 c4heoA_
6.7
31
PDB header: viral proteinChain: A: PDB Molecule: phosphoprotein;PDBTitle: hendra virus phosphoprotein c terminal domain
17 c2ogdB_
6.6
14
PDB header: transferaseChain: B: PDB Molecule: farnesyl pyrophosphate synthase;PDBTitle: t. brucei farnesyl diphosphate synthase complexed with bisphosphonate2 bph-527
18 c3ijpA_
6.4
25
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
19 d1vm6a2
6.2
26
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
20 c2v6qB_
6.2
33
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of a bhrf-1 : bim bh3 complex
21 c2wh6B_
not modelled
6.1
33
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of anti-apoptotic bhrf1 in complex with the bim bh32 domain
22 c5vwzD_
not modelled
6.0
33
PDB header: apoptosisChain: D: PDB Molecule: bcl-2-like protein 11;PDBTitle: bak in complex with bim-h3pc
23 c5vwzB_
not modelled
6.0
33
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: bak in complex with bim-h3pc
24 c4nlbA_
not modelled
5.9
32
PDB header: hydrolaseChain: A: PDB Molecule: ribosomal rna processing protein 6;PDBTitle: crystal structure of the catalytic core of rrp6 from trypanosoma2 brucei
25 c3fdlB_
not modelled
5.9
33
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: bim bh3 peptide in complex with bcl-xl
26 c4ywjB_
not modelled
5.7
25
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate reductase (htpa2 reductase) from pseudomonas aeruginosa
27 c4a1bB_
not modelled
5.6
38
PDB header: ribosomeChain: B: PDB Molecule: rpl39;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 3.
28 c2ndjA_
not modelled
5.5
16
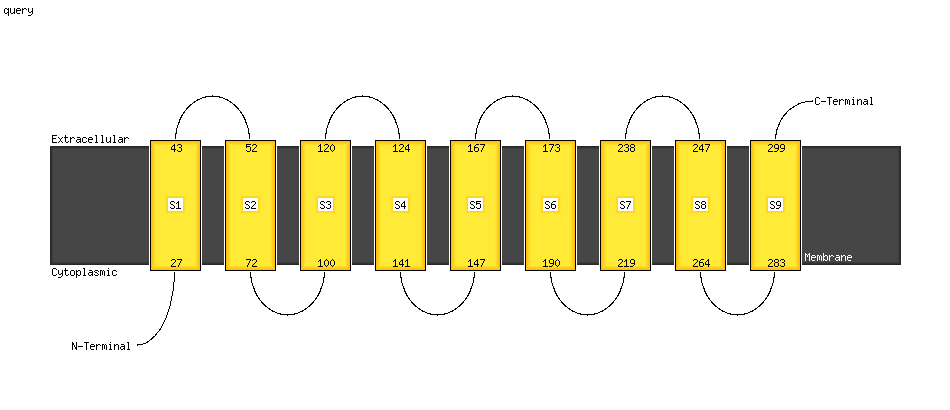
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 3;PDBTitle: structural basis for kcne3 and estrogen modulation of the kcnq12 channel
29 c5k4bA_
not modelled
5.4
25
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 3 subunit d;PDBTitle: structure of eukaryotic translation initiation factor 3 subunit d2 (eif3d) cap binding domain from nasonia vitripennis, crystal form 1
30 c2mpnB_
not modelled
5.2
12
PDB header: membrane proteinChain: B: PDB Molecule: inner membrane protein ygap;PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
31 c2hbkA_
not modelled
5.2
19
PDB header: hydrolase, gene regulationChain: A: PDB Molecule: exosome complex exonuclease rrp6;PDBTitle: structure of the yeast nuclear exosome component, rrp6p, reveals an2 interplay between the active site and the hrdc domain; protein in3 complex with mn
32 c6nplA_
not modelled
5.2
12
PDB header: membrane proteinChain: A: PDB Molecule: solute carrier family 12 (sodium/potassium/chloridePDBTitle: cryo-em structure of nkcc1
33 c3izso_
not modelled
5.2
38
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein rpl28 (l15p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
34 c6g2jY_
not modelled
5.2
7
PDB header: oxidoreductaseChain: Y: PDB Molecule: mcg5603;PDBTitle: mouse mitochondrial complex i in the active state
35 c3ukwC_
not modelled
5.2
40
PDB header: protein transport/inhibitorChain: C: PDB Molecule: bimax1 peptide;PDBTitle: mouse importin alpha: bimax1 peptide complex
36 c2zkr3_
not modelled
5.1
50
PDB header: ribosomal protein/rnaChain: 3: PDB Molecule: 60s ribosomal protein l39e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
37 c3qy9C_
not modelled
5.0
32
PDB header: oxidoreductaseChain: C: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: the crystal structure of dihydrodipicolinate reductase from2 staphylococcus aureus