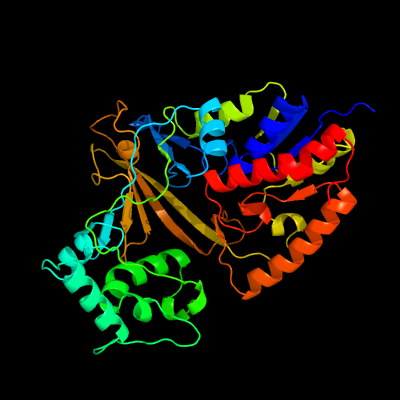
| 1 | c1v0jB_
|
|
 |
100.0 |
98 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: udp-galactopyranose mutase from mycobacterium tuberculosis
|
|
|
|
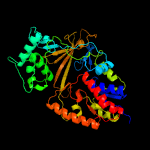
| 2 | c4mo2A_
|
|
 |
100.0 |
44 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: crystal structure of udp-n-acetylgalactopyranose mutase from2 campylobacter jejuni
|
|
|
|
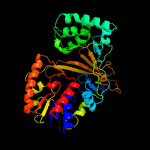
| 3 | c1i8tB_
|
|
 |
100.0 |
44 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: strcuture of udp-galactopyranose mutase from e.coli
|
|
|
|
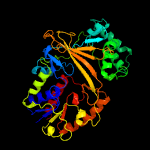
| 4 | c2bi8A_
|
|
 |
100.0 |
41 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: udp-galactopyranose mutase from klebsiella pneumoniae with reduced fad
|
|
|
|
| 5 | c3hdqI_
|
|
 |
100.0 |
40 |
PDB header:isomerase
Chain: I: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: crystal structure of udp-galactopyranose mutase (oxidized2 form) in complex with substrate
|
|
|
|
| 6 | c3uteB_
|
|
 |
100.0 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: crystal structure of aspergillus fumigatus udp galactopyranose mutase2 sulfate complex
|
|
|
|
| 7 | d2bi7a1
|
|
 |
100.0 |
41 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:UDP-galactopyranose mutase, N-terminal domain |
|
|
|
| 8 | d1i8ta1
|
|
 |
100.0 |
43 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:UDP-galactopyranose mutase, N-terminal domain |
|
|
|
| 9 | c4dshB_
|
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: crystal structure of reduced udp-galactopyranose mutase
|
|
|
|
| 10 | c2ivdA_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: structure of protoporphyrinogen oxidase from myxococcus2 xanthus with acifluorfen
|
|
|
|
| 11 | c1s3bB_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine oxidase [flavin-containing] b;
PDBTitle: crystal structure of maob in complex with n-methyl-n-2 propargyl-1(r)-aminoindan
|
|
|
|
| 12 | c5ttkB_
|
|
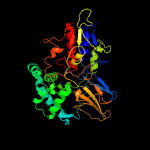 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine oxidase;
PDBTitle: crystal structure of selenomethionine-incorporated nicotine2 oxidoreductase from pseudomonas putida
|
|
|
|
| 13 | c3nksA_
|
|
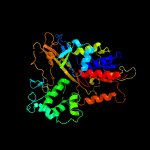 |
100.0 |
14 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: structure of human protoporphyrinogen ix oxidase
|
|
|
|
| 14 | c3lovA_
|
|
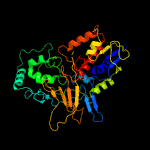 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: crystal structure of putative protoporphyrinogen oxidase2 (yp_001813199.1) from exiguobacterium sp. 255-15 at 2.06 a resolution
|
|
|
|
| 15 | c3i6dA_
|
|
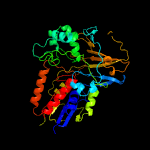 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase;
PDBTitle: crystal structure of ppo from bacillus subtilis with af
|
|
|
|
| 16 | c4gutA_
|
|
 |
100.0 |
16 |
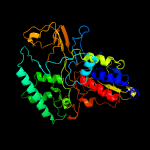
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific histone demethylase 1b;
PDBTitle: crystal structure of lsd2-npac
|
|
|
|
| 17 | c4i58A_
|
|
 |
100.0 |
14 |
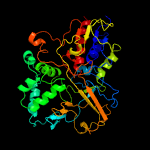
PDB header:oxidoreductase
Chain: A: PDB Molecule:cyclohexylamine oxidase;
PDBTitle: cyclohexylamine oxidase from brevibacterium oxydans ih-35a
|
|
|
|
| 18 | c6cr0A_
|
|
 |
100.0 |
15 |
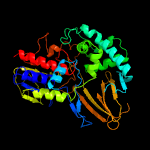
PDB header:oxidoreductase
Chain: A: PDB Molecule:(s)-6-hydroxynicotine oxidase;
PDBTitle: 1.55 a resolution structure of (s)-6-hydroxynicotine oxidase from2 shinella hzn7
|
|
|
|
| 19 | c1f8sA_
|
|
 |
100.0 |
13 |
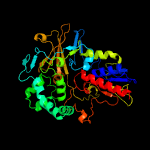
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-amino acid oxidase;
PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
|
|
|
|
| 20 | c3x0vA_
|
|
 |
100.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lysine oxidase;
PDBTitle: structure of l-lysine oxidase
|
|
|
|
| 21 | c1sezA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protoporphyrinogen oxidase, mitochondrial;
PDBTitle: crystal structure of protoporphyrinogen ix oxidase
|
|
|
| 22 | c3k7tB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-hydroxy-l-nicotine oxidase;
PDBTitle: crystal structure of apo-form 6-hydroxy-l-nicotine oxidase, crystal2 form p3121
|
|
|
| 23 | c5mogB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phytoene dehydrogenase, chloroplastic/chromoplastic;
PDBTitle: oryza sativa phytoene desaturase inhibited by norflurazon
|
|
|
| 24 | c3rhaA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putrescine oxidase;
PDBTitle: the crystal structure of oxidoreductase from arthrobacter aurescens
|
|
|
| 25 | c4iv9B_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:tryptophan 2-monooxygenase;
PDBTitle: structure of the flavoprotein tryptophan-2-monooxygenase
|
|
|
| 26 | c6fjhB_ |
|
not modelled |
100.0 |
15 |
PDB header:flavoprotein
Chain: B: PDB Molecule:lkce;
PDBTitle: crystal structure of the seleniated lkce from streptomyces rochei
|
|
|
| 27 | c2b9yA_ |
|
not modelled |
100.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aminooxidase;
PDBTitle: crystal structure of cla-producing fatty acid isomerase2 from p. acnes
|
|
|
| 28 | c2vvlD_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:monoamine oxidase n;
PDBTitle: the structure of mao-n-d3, a variant of monoamine oxidase2 from aspergillus niger.
|
|
|
| 29 | c3we0A_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-amino acid oxidase/monooxygenase;
PDBTitle: l-amino acid oxidase/monooxygenase from pseudomonas sp. aiu 813
|
|
|
| 30 | c2yg4B_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putrescine oxidase;
PDBTitle: structure-based redesign of cofactor binding in putrescine2 oxidase: wild type bound to putrescine
|
|
|
| 31 | c5mbxA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peroxisomal n(1)-acetyl-spermine/spermidine oxidase;
PDBTitle: crystal structure of reduced murine n1-acetylpolyamine oxidase in2 complex with n1-acetylspermine
|
|
|
| 32 | d1o5wa1 |
|
not modelled |
100.0 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 33 | c5g3sB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-tryptophan oxidase vioa;
PDBTitle: the structure of the l-tryptophan oxidase vioa from chromobacterium2 violaceum - samarium derivative
|
|
|
| 34 | c2hkoA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: crystal structure of lsd1
|
|
|
| 35 | c2v1dA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase/repressor
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
|
|
|
| 36 | c3bnuA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyamine oxidase fms1;
PDBTitle: crystal structure of polyamine oxidase fms1 from2 saccharomyces cerevisiae in complex with bis-(3s,3's)-3 methylated spermine
|
|
|
| 37 | c2xagA_ |
|
not modelled |
100.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: crystal structure of lsd1-corest in complex with para-bromo-2 (-)-trans-2-phenylcyclopropyl-1-amine
|
|
|
| 38 | c2yr6A_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pro-enzyme of l-phenylalanine oxidase;
PDBTitle: crystal structure of l-phenylalanine oxidase from psuedomonas sp.p501
|
|
|
| 39 | c2jb1B_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-amino acid oxidase;
PDBTitle: the l-amino acid oxidase from rhodococcus opacus in complex2 with l-alanine
|
|
|
| 40 | c1ltxR_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase/protein binding
Chain: R: PDB Molecule:rab escort protein 1;
PDBTitle: structure of rab escort protein-1 in complex with rab geranylgeranyl2 transferase and isoprenoid
|
|
|
| 41 | d2v5za1 |
|
not modelled |
100.0 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 42 | c1h83A_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyamine oxidase;
PDBTitle: structure of polyamine oxidase in complex with2 1,8-diaminooctane
|
|
|
| 43 | c3ka7A_ |
|
not modelled |
100.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of an oxidoreductase from methanosarcina2 mazei. northeast structural genomics consortium target id3 mar208
|
|
|
| 44 | c4repA_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase, flavoprotein
Chain: A: PDB Molecule:gamma-carotene desaturase;
PDBTitle: crystal structure of gamma-carotenoid desaturase
|
|
|
| 45 | d2dw4a2 |
|
not modelled |
100.0 |
13 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 46 | c6c87A_ |
|
not modelled |
100.0 |
12 |
PDB header:protein transport
Chain: A: PDB Molecule:rab gdp dissociation inhibitor alpha;
PDBTitle: crystal structure of rab gdp dissociation inhibitor alpha from2 naegleria fowleri
|
|
|
| 47 | d1reoa1 |
|
not modelled |
99.9 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 48 | d2iida1 |
|
not modelled |
99.9 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 49 | c3cpiH_ |
|
not modelled |
99.9 |
14 |
PDB header:protein transport
Chain: H: PDB Molecule:rab gdp-dissociation inhibitor;
PDBTitle: crystal structure of yeast rab-gdi
|
|
|
| 50 | c3nrnA_ |
|
not modelled |
99.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pf1083;
PDBTitle: crystal structure of pf1083 protein from pyrococcus furiosus,2 northeast structural genomics consortium target pfr223
|
|
|
| 51 | d1b5qa1 |
|
not modelled |
99.9 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 52 | c4dgkA_ |
|
not modelled |
99.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phytoene dehydrogenase;
PDBTitle: crystal structure of phytoene desaturase crti from pantoea ananatis
|
|
|
| 53 | c1gndA_ |
|
not modelled |
99.9 |
15 |
PDB header:gtpase activation
Chain: A: PDB Molecule:guanine nucleotide dissociation inhibitor;
PDBTitle: guanine nucleotide dissociation inhibitor, alpha-isoform
|
|
|
| 54 | d2ivda1 |
|
not modelled |
99.9 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 55 | c3p1wA_ |
|
not modelled |
99.9 |
12 |
PDB header:protein transport
Chain: A: PDB Molecule:rabgdi protein;
PDBTitle: crystal structure of rab gdi from plasmodium falciparum, pfl2060c
|
|
|
| 56 | c2e1mA_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-glutamate oxidase;
PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
|
|
|
| 57 | d1d5ta1 |
|
not modelled |
99.9 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
|
|
| 58 | d1seza1 |
|
not modelled |
99.9 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 59 | d2bcgg1 |
|
not modelled |
99.9 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
|
|
| 60 | d1vg0a1 |
|
not modelled |
99.9 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
|
|
| 61 | c3qj4A_ |
|
not modelled |
99.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:renalase;
PDBTitle: crystal structure of human renalase (isoform 1)
|
|
|
| 62 | c4ia6B_ |
|
not modelled |
99.7 |
13 |
PDB header:immune system
Chain: B: PDB Molecule:myosin-crossreactive antigen;
PDBTitle: hydratase from lactobacillus acidophilus in a ligand bound form (la2 lah)
|
|
|
| 63 | c1yvvB_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine oxidase, flavin-containing;
PDBTitle: x-ray structurure of p. syringae q888a4 oxidoreductase at2 resolution 2.5a. northeast structural genomics consortium3 target psr10.
|
|
|
| 64 | c1ps9A_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-dienoyl2 coa reductase
|
|
|
| 65 | d2gqfa1 |
|
not modelled |
99.5 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
|
|
| 66 | d2gmha1 |
|
not modelled |
99.5 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 67 | d2gf3a1 |
|
not modelled |
99.5 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 68 | c3i3lA_ |
|
not modelled |
99.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkylhalidase cmls;
PDBTitle: crystal structure of cmls, a flavin-dependent halogenase
|
|
|
| 69 | c4xwzA_ |
|
not modelled |
99.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl amine:oxygen oxidoreductase;
PDBTitle: the crystal structure of fructosyl amine: oxygen oxidoreductase2 (amadoriase i) from aspergillus fumigatus in complex with the3 substrate fructosyl lysine
|
|
|
| 70 | c2olnA_ |
|
not modelled |
99.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nikd protein;
PDBTitle: nikd, an unusual amino acid oxidase essential for2 nikkomycin biosynthesis: closed form at 1.15 a resolution
|
|
|
| 71 | c3dmeB_ |
|
not modelled |
99.4 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved exported protein;
PDBTitle: crystal structure of conserved exported protein from bordetella2 pertussis. northeast structural genomics target ber141
|
|
|
| 72 | c3nixF_ |
|
not modelled |
99.3 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:flavoprotein/dehydrogenase;
PDBTitle: crystal structure of flavoprotein/dehydrogenase from cytophaga2 hutchinsonii. northeast structural genomics consortium target chr43.
|
|
|
| 73 | c6bwtD_ |
|
not modelled |
99.3 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:thioredoxin reductase;
PDBTitle: 2.45 angstrom resolution crystal structure thioredoxin reductase from2 francisella tularensis.
|
|
|
| 74 | c1v59B_ |
|
not modelled |
99.3 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: crystal structure of yeast lipoamide dehydrogenase2 complexed with nad+
|
|
|
| 75 | c3axbA_ |
|
not modelled |
99.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of a dye-linked l-proline dehydrogenase from the aerobic2 hyperthermophilic archaeon, aeropyrum pernix
|
|
|
| 76 | c2w0hA_ |
|
not modelled |
99.3 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trypanothione reductase;
PDBTitle: x ray structure of leishmania infantum trypanothione2 reductase in complex with antimony and nadph
|
|
|
| 77 | c2qa2A_ |
|
not modelled |
99.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyketide oxygenase cabe;
PDBTitle: crystal structure of cabe, an aromatic hydroxylase from angucycline2 biosynthesis, determined to 2.7 a resolution
|
|
|
| 78 | c2rghA_ |
|
not modelled |
99.3 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
|
|
| 79 | c1tytA_ |
|
not modelled |
99.3 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trypanothione reductase, oxidized form;
PDBTitle: crystal and molecular structure of crithidia fasciculata2 trypanothione reductase at 2.6 angstroms resolution
|
|
|
| 80 | c2gqfA_ |
|
not modelled |
99.2 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hi0933;
PDBTitle: crystal structure of flavoprotein hi0933 from haemophilus influenzae2 rd
|
|
|
| 81 | c5v36A_ |
|
not modelled |
99.2 |
31 |
PDB header:hydrolase,oxidoreductase
Chain: A: PDB Molecule:glutathione reductase;
PDBTitle: 1.88 angstrom resolution crystal structure of glutathione reductase2 from streptococcus mutans ua159 in complex with fad
|
|
|
| 82 | c1dxlC_ |
|
not modelled |
99.2 |
26 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: dihydrolipoamide dehydrogenase of glycine decarboxylase2 from pisum sativum
|
|
|
| 83 | c5xgvB_ |
|
not modelled |
99.2 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyre3;
PDBTitle: the structure of diels-alderase pyre3 in the biosynthetic pathway of2 pyrroindomycins
|
|
|
| 84 | d1ryia1 |
|
not modelled |
99.2 |
27 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 85 | c2rgoA_ |
|
not modelled |
99.2 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
|
|
| 86 | c2cduB_ |
|
not modelled |
99.2 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph oxidase;
PDBTitle: the crystal structure of water-forming nad(p)h oxidase from2 lactobacillus sanfranciscensis
|
|
|
| 87 | d1v59a1 |
|
not modelled |
99.2 |
27 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 88 | c1ojtA_ |
|
not modelled |
99.2 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:surface protein;
PDBTitle: structure of dihydrolipoamide dehydrogenase
|
|
|
| 89 | c4cnjD_ |
|
not modelled |
99.2 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:l-amino acid oxidase;
PDBTitle: l-aminoacetone oxidase from streptococcus oligofermentans2 belongs to a new 3-domain family of bacterial flavoproteins
|
|
|
| 90 | c3da1A_ |
|
not modelled |
99.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
|
|
|
| 91 | c2qaeA_ |
|
not modelled |
99.2 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure analysis of trypanosoma cruzi lipoamide2 dehydrogenase
|
|
|
| 92 | c4usrA_ |
|
not modelled |
99.2 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: structure of flavin-containing monooxygenase from2 pseudomonas stutzeri nf13
|
|
|
| 93 | c5hxwF_ |
|
not modelled |
99.2 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:l-amino acid deaminase;
PDBTitle: l-amino acid deaminase from proteus vulgaris
|
|
|
| 94 | c2i0zA_ |
|
not modelled |
99.2 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-utilizing dehydrogenases;
PDBTitle: crystal structure of a fad binding protein from bacillus2 cereus, a putative nad(fad)-utilizing dehydrogenases
|
|
|
| 95 | c2c3dB_ |
|
not modelled |
99.2 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxopropyl-com reductase;
PDBTitle: 2.15 angstrom crystal structure of 2-ketopropyl coenzyme m2 oxidoreductase carboxylase with a coenzyme m disulfide3 bound at the active site
|
|
|
| 96 | c2qa1A_ |
|
not modelled |
99.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyketide oxygenase pgae;
PDBTitle: crystal structure of pgae, an aromatic hydroxylase involved in2 angucycline biosynthesis
|
|
|
| 97 | c2eq7B_ |
|
not modelled |
99.2 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxoglutarate dehydrogenase e3 component;
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdo
|
|
|
| 98 | c5fjnB_ |
|
not modelled |
99.2 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:l-amino acid deaminase;
PDBTitle: structure of l-amino acid deaminase from proteus myxofaciens2 in complex with anthranilate
|
|
|
| 99 | c3l8kB_ |
|
not modelled |
99.1 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoyl dehydrogenase from sulfolobus2 solfataricus
|
|
|
| 100 | c3urhB_ |
|
not modelled |
99.1 |
41 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoamide dehydrogenase from2 sinorhizobium meliloti 1021
|
|
|
| 101 | c4b1bB_ |
|
not modelled |
99.1 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of plasmodium falciparum oxidised2 thioredoxin reductase at 2.9 angstrom
|
|
|
| 102 | c1ndaD_ |
|
not modelled |
99.1 |
40 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:trypanothione oxidoreductase;
PDBTitle: the structure of trypanosoma cruzi trypanothione reductase in the2 oxidized and nadph reduced state
|
|
|
| 103 | c5glgA_ |
|
not modelled |
99.1 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase 2;
PDBTitle: the novel function of osm1 under anaerobic condition in the er was2 revealed by crystal structure of osm1, a soluble fumarate reductase3 in yeast
|
|
|
| 104 | c4at2A_ |
|
not modelled |
99.1 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-ketosteroid-delta4-5alpha-dehydrogenase;
PDBTitle: the crystal structure of 3-ketosteroid-delta4-(5alpha)-2 dehydrogenase from rhodococcus jostii rha1 in complex3 with 4-androstene-3,17- dione
|
|
|
| 105 | c1lpfB_ |
|
not modelled |
99.1 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: three-dimensional structure of lipoamide dehydrogenase from2 pseudomonas fluorescens at 2.8 angstroms resolution.3 analysis of redox and thermostability properties
|
|
|
| 106 | c1ryiB_ |
|
not modelled |
99.1 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycine oxidase;
PDBTitle: structure of glycine oxidase with bound inhibitor glycolate
|
|
|
| 107 | c6du7C_ |
|
not modelled |
99.1 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glutathione reductase;
PDBTitle: glutathione reductase from streptococcus pneumoniae
|
|
|
| 108 | c1zmcG_ |
|
not modelled |
99.1 |
29 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of human dihydrolipoamide dehydrogenase2 complexed to nad+
|
|
|
| 109 | c2gmhA_ |
|
not modelled |
99.1 |
37 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:electron transfer flavoprotein-ubiquinone
PDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
|
|
|
| 110 | d1dxla1 |
|
not modelled |
99.1 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 111 | d3lada1 |
|
not modelled |
99.1 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 112 | d2i0za1 |
|
not modelled |
99.1 |
31 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
|
|
| 113 | c6aonB_ |
|
not modelled |
99.1 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: 1.72 angstrom resolution crystal structure of 2-oxoglutarate2 dehydrogenase complex subunit dihydrolipoamide dehydrogenase from3 bordetella pertussis in complex with fad
|
|
|
| 114 | c3rp7A_ |
|
not modelled |
99.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavoprotein monooxygenase;
PDBTitle: crystal structure of klebsiella pneumoniae hpxo complexed with fad and2 uric acid
|
|
|
| 115 | c2r4jA_ |
|
not modelled |
99.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aerobic glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of escherichia coli semet substituted glycerol-3-2 phosphate dehydrogenase in complex with dhap
|
|
|
| 116 | c1pj6A_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n,n-dimethylglycine oxidase;
PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
|
|
|
| 117 | c3nlcA_ |
|
not modelled |
99.1 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vp0956;
PDBTitle: crystal structure of the vp0956 protein from vibrio parahaemolyticus.2 northeast structural genomics consortium target vpr147
|
|
|
| 118 | c6de6B_ |
|
not modelled |
99.1 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: 2.1 a resolution structure of histamine dehydrogenase from rhizobium2 sp. 4-9
|
|
|
| 119 | c4x9mA_ |
|
not modelled |
99.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-alpha-glycerophosphate oxidase;
PDBTitle: oxidized l-alpha-glycerophosphate oxidase from mycoplasma pneumoniae2 with fad bound
|
|
|
| 120 | c1zkqA_ |
|
not modelled |
99.1 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2, mitochondrial;
PDBTitle: crystal structure of mouse thioredoxin reductase type 2
|
|
|