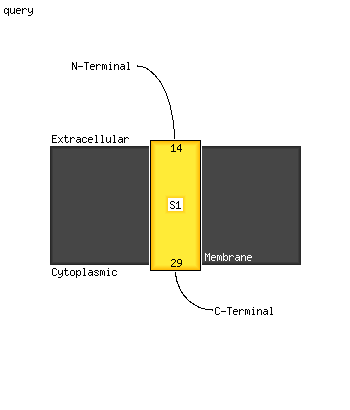
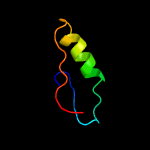
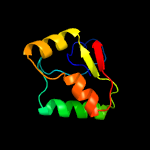
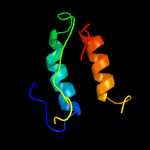
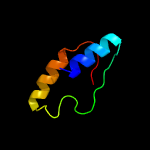
1 c5kymA_
100.0
23
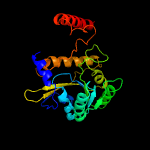
PDB header: transferaseChain: A: PDB Molecule: 1-acyl-sn-glycerol-3-phosphate acyltransferase;PDBTitle: crystal structure of the 1-acyl-sn-glycerophosphate (lpa)2 acyltransferase, plsc, from thermotoga maritima
2 c5f34A_
99.8
14
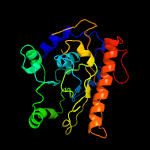
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol mannoside acyltransferase;PDBTitle: crystal structure of membrane associated pata from mycobacterium2 smegmatis in complex with s-hexadecyl coenzyme a - p21 space group
3 c5knkB_
99.7
17
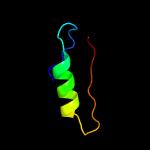
PDB header: transferaseChain: B: PDB Molecule: lipid a biosynthesis lauroyl acyltransferase;PDBTitle: lipid a secondary acyltransferase lpxm from acinetobacter baumannii2 with catalytic residue substitution (e127a)
4 d1iuqa_
99.1
14
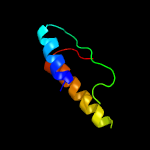
Fold: Glycerol-3-phosphate (1)-acyltransferaseSuperfamily: Glycerol-3-phosphate (1)-acyltransferaseFamily: Glycerol-3-phosphate (1)-acyltransferase
5 c6g2dC_
35.5
12
PDB header: ligaseChain: C: PDB Molecule: acetyl-coa carboxylase 1;PDBTitle: citrate-induced acetyl-coa carboxylase (acc-cit) filament at 5.4 a2 resolution
6 c4mxnB_
31.7
8
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative glycosyl hydrolase (parmer_00599) from2 parabacteroides merdae atcc 43184 at 1.95 a resolution
7 c3hkxA_
22.7
14
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
8 c2ov3A_
21.9
13
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein component of an abc type zincPDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc bound
9 c3zppA_
20.3
19
PDB header: cell adhesionChain: A: PDB Molecule: cell wall surface anchor family protein;PDBTitle: structure of the streptococcus pneumoniae surface protein2 and adhesin pfba
10 c4cyyA_
19.4
12
PDB header: hydrolaseChain: A: PDB Molecule: pantetheinase;PDBTitle: the structure of vanin-1: defining the link between metabolic disease,2 oxidative stress and inflammation
11 c3jurA_
18.5
16
PDB header: hydrolaseChain: A: PDB Molecule: exo-poly-alpha-d-galacturonosidase;PDBTitle: the crystal structure of a hyperthermoactive exopolygalacturonase from2 thermotoga maritima
12 c5olpB_
17.3
19
PDB header: hydrolaseChain: B: PDB Molecule: pectate lyase;PDBTitle: galacturonidase
13 d1pq4a_
17.1
12
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
14 c2e11B_
16.0
16
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase;PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
15 c5khaA_
15.1
12
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad+ synthetase;PDBTitle: structure of glutamine-dependent nad+ synthetase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp)
16 c1r0lD_
15.0
15
PDB header: oxidoreductaseChain: D: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis2 in complex with nadph
17 c2ogwB_
14.5
18
PDB header: transport proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znuaPDBTitle: structure of abc type zinc transporter from e. coli
18 c5uyvA_
14.0
18
PDB header: metal transportChain: A: PDB Molecule: periplasmic chelated iron-binding protein yfea;PDBTitle: yfea ancillary sites that do not co-load with site 2
19 c4hnvB_
12.9
13
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of r54e mutant of s. aureus pyruvate carboxylase
20 d1v7za_
12.5
17
Fold: CreatininaseSuperfamily: CreatininaseFamily: Creatininase
21 c6r6kB_
not modelled
12.0
17
PDB header: protein transportChain: B: PDB Molecule: abc transporter substrate-binding protein;PDBTitle: structure of a fpvc mutant from pseudomonas aeruginosa
22 c3a14B_
not modelled
11.7
17
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
23 d2ffca1
not modelled
11.6
14
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
24 c2pyhB_
not modelled
11.0
16
PDB header: isomeraseChain: B: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: azotobacter vinelandii mannuronan c-5 epimerase alge4 a-module2 complexed with mannuronan trisaccharide
25 c5n6mA_
not modelled
10.8
12
PDB header: membrane proteinChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: structure of the membrane integral lipoprotein n-acyltransferase lnt2 from p. aeruginosa
26 c5vrhA_
not modelled
10.7
9
PDB header: transferaseChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
27 c3va7A_
not modelled
10.4
21
PDB header: ligaseChain: A: PDB Molecule: klla0e08119p;PDBTitle: crystal structure of the kluyveromyces lactis urea carboxylase
28 c3w7bB_
not modelled
10.4
17
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase from thermus2 thermophilus hb8
29 c3bg5B_
not modelled
10.0
13
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of staphylococcus aureus pyruvate carboxylase
30 c2o1eB_
not modelled
9.8
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ycdh;PDBTitle: crystal structure of the metal-dependent lipoprotein ycdh2 from bacillus subtilis, northeast structural genomics3 target sr583
31 c2ps3A_
not modelled
9.8
18
PDB header: metal transportChain: A: PDB Molecule: high-affinity zinc uptake system protein znua;PDBTitle: structure and metal binding properties of znua, a periplasmic zinc2 transporter from escherichia coli
32 c4cl2A_
not modelled
9.7
15
PDB header: transport proteinChain: A: PDB Molecule: periplasmic solute binding protein;PDBTitle: structure of periplasmic metal binding protein from candidatus2 liberibacter asiaticus
33 c3ve9B_
not modelled
9.4
10
PDB header: lyaseChain: B: PDB Molecule: orotidine-5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 metallosphaera sedula
34 c1emsB_
not modelled
9.1
10
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
35 d1kl7a_
not modelled
8.7
15
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
36 c2uveA_
not modelled
8.7
15
PDB header: hydrolaseChain: A: PDB Molecule: exopolygalacturonase;PDBTitle: structure of yersinia enterocolitica family 282 exopolygalacturonase
37 d1hrua_
not modelled
8.5
29
Fold: YrdC/RibBSuperfamily: YrdC/RibBFamily: YrdC-like
38 d2q8za1
not modelled
8.4
22
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
39 d1o59a2
not modelled
8.2
18
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Allantoicase repeat
40 c4imyI_
not modelled
8.2
9
PDB header: transferaseChain: I: PDB Molecule: af4/fmr2 family member 4;PDBTitle: the aff4 scaffold binds human p-tefb adjacent to hiv tat
41 c4imyG_
not modelled
8.2
9
PDB header: transferaseChain: G: PDB Molecule: af4/fmr2 family member 4;PDBTitle: the aff4 scaffold binds human p-tefb adjacent to hiv tat
42 c4imyH_
not modelled
8.1
9
PDB header: transferaseChain: H: PDB Molecule: af4/fmr2 family member 4;PDBTitle: the aff4 scaffold binds human p-tefb adjacent to hiv tat
43 d1lg7a_
not modelled
8.1
22
Fold: VSV matrix proteinSuperfamily: VSV matrix proteinFamily: VSV matrix protein
44 d1o59a1
not modelled
7.9
27
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Allantoicase repeat
45 d1aopa2
not modelled
7.5
21
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 3
46 c3eqnB_
not modelled
7.4
14
PDB header: hydrolaseChain: B: PDB Molecule: glucan 1,3-beta-glucosidase;PDBTitle: crystal structure of beta-1,3-glucanase from phanerochaete2 chrysosporium (lam55a)
47 c4xrvB_
not modelled
7.4
19
PDB header: metal binding proteinChain: B: PDB Molecule: periplasmic solute binding protein;PDBTitle: structure of a zn abc transporter substrate binding protein from2 paracoccus denitrificans
48 c2eghA_
not modelled
6.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
49 d2fdsa1
not modelled
6.7
8
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
50 c2fdsA_
not modelled
6.7
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: orotidine-monophosphate-decarboxylase;PDBTitle: crystal structure of plasmodium berghei orotidine 5'-monophosphate2 decarboxylase (ortholog of plasmodium falciparum pf10_0225)
51 d1q0qa2
not modelled
6.3
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
52 d1w98b1
not modelled
6.2
15
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin
53 d1rmga_
not modelled
6.1
8
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase
54 c3mfqB_
not modelled
6.0
14
PDB header: metal binding proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znua;PDBTitle: a glance into the metal binding specificity of troa: where elaborate2 behaviors occur in the active center
55 c2zo7A_
not modelled
5.4
31
PDB header: luminescent proteinChain: A: PDB Molecule: cyan/green-emitting gfp-like protein, kusabira-cyan mutantPDBTitle: crystal structure of a kusabira-cyan mutant (kcy-r1), a cyan/green-2 emitting gfp-like protein
56 c4qskB_
not modelled
5.3
19
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of l. monocytogenes pyruvate carboxylase in complex2 with cyclic-di-amp
57 c3obiC_
not modelled
5.3
11
PDB header: hydrolaseChain: C: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
58 d1xvla1
not modelled
5.2
13
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like