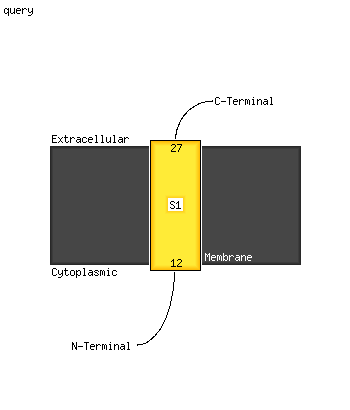
| 1 |
|
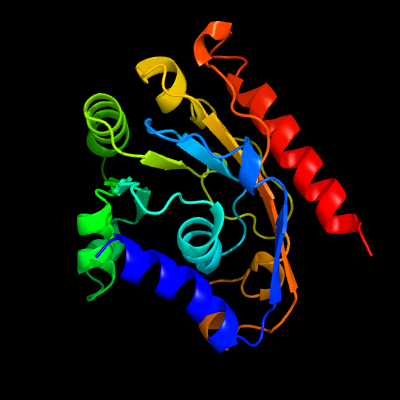
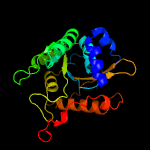
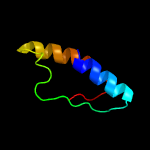
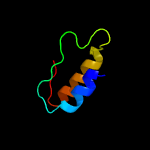
PDB 5kym chain A
Region: 5 - 207
Aligned: 193
Modelled: 203
Confidence: 100.0%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:1-acyl-sn-glycerol-3-phosphate acyltransferase;
PDBTitle: crystal structure of the 1-acyl-sn-glycerophosphate (lpa)2 acyltransferase, plsc, from thermotoga maritima
Phyre2
| 2 |
|
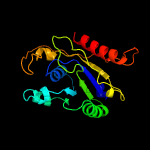
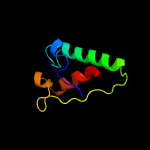
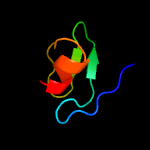
PDB 5f34 chain A
Region: 1 - 207
Aligned: 192
Modelled: 192
Confidence: 99.8%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol mannoside acyltransferase;
PDBTitle: crystal structure of membrane associated pata from mycobacterium2 smegmatis in complex with s-hexadecyl coenzyme a - p21 space group
Phyre2
| 3 |
|
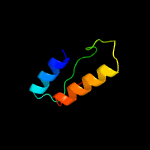
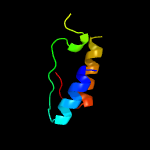
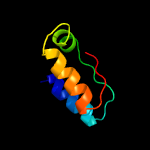
PDB 5knk chain B
Region: 1 - 228
Aligned: 205
Modelled: 212
Confidence: 99.6%
Identity: 14%
PDB header:transferase
Chain: B: PDB Molecule:lipid a biosynthesis lauroyl acyltransferase;
PDBTitle: lipid a secondary acyltransferase lpxm from acinetobacter baumannii2 with catalytic residue substitution (e127a)
Phyre2
| 4 |
|
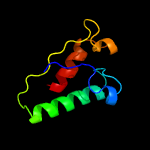
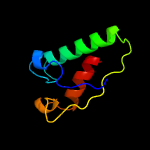
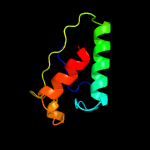
PDB 1iuq chain A
Region: 30 - 205
Aligned: 171
Modelled: 176
Confidence: 99.0%
Identity: 17%
Fold: Glycerol-3-phosphate (1)-acyltransferase
Superfamily: Glycerol-3-phosphate (1)-acyltransferase
Family: Glycerol-3-phosphate (1)-acyltransferase
Phyre2
| 5 |
|
PDB 4cyy chain A
Region: 96 - 145
Aligned: 50
Modelled: 50
Confidence: 18.4%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:pantetheinase;
PDBTitle: the structure of vanin-1: defining the link between metabolic disease,2 oxidative stress and inflammation
Phyre2
| 6 |
|
PDB 1l8n chain A domain 1
Region: 171 - 244
Aligned: 74
Modelled: 74
Confidence: 13.0%
Identity: 11%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: alpha-D-glucuronidase/Hyaluronidase catalytic domain
Phyre2
| 7 |
|
PDB 5kha chain A
Region: 96 - 145
Aligned: 50
Modelled: 50
Confidence: 11.6%
Identity: 18%
PDB header:ligase
Chain: A: PDB Molecule:glutamine-dependent nad+ synthetase;
PDBTitle: structure of glutamine-dependent nad+ synthetase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp)
Phyre2
| 8 |
|
PDB 1h41 chain A domain 1
Region: 171 - 244
Aligned: 74
Modelled: 74
Confidence: 11.5%
Identity: 11%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: alpha-D-glucuronidase/Hyaluronidase catalytic domain
Phyre2
| 9 |
|
PDB 3hkx chain A
Region: 96 - 145
Aligned: 50
Modelled: 50
Confidence: 10.5%
Identity: 12%
PDB header:hydrolase
Chain: A: PDB Molecule:amidase;
PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
Phyre2
| 10 |
|
PDB 1mqr chain A
Region: 171 - 244
Aligned: 74
Modelled: 74
Confidence: 9.8%
Identity: 11%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-d-glucuronidase;
PDBTitle: the crystal structure of alpha-d-glucuronidase (e386q) from bacillus2 stearothermophilus t-6
Phyre2
| 11 |
|
PDB 5vrh chain A
Region: 97 - 145
Aligned: 49
Modelled: 49
Confidence: 9.5%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:apolipoprotein n-acyltransferase;
PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
Phyre2
| 12 |
|
PDB 1wx4 chain B
Region: 21 - 56
Aligned: 36
Modelled: 36
Confidence: 8.8%
Identity: 22%
PDB header:oxidoreductase/metal transport
Chain: B: PDB Molecule:melc;
PDBTitle: crystal structure of the oxy-form of the copper-bound streptomyces2 castaneoglobisporus tyrosinase complexed with a caddie protein3 prepared by the addition of dithiothreitol
Phyre2
| 13 |
|
PDB 1ems chain B
Region: 96 - 146
Aligned: 51
Modelled: 51
Confidence: 8.8%
Identity: 8%
PDB header:antitumor protein
Chain: B: PDB Molecule:nit-fragile histidine triad fusion protein;
PDBTitle: crystal structure of the c. elegans nitfhit protein
Phyre2
| 14 |
|
PDB 1gqk chain B
Region: 171 - 244
Aligned: 74
Modelled: 74
Confidence: 8.7%
Identity: 11%
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-d-glucuronidase;
PDBTitle: structure of pseudomonas cellulosa alpha-d-glucuronidase complexed2 with glucuronic acid
Phyre2
| 15 |
|
PDB 1uf5 chain A
Region: 96 - 145
Aligned: 50
Modelled: 49
Confidence: 7.8%
Identity: 16%
Fold: Carbon-nitrogen hydrolase
Superfamily: Carbon-nitrogen hydrolase
Family: Carbamilase
Phyre2
| 16 |
|
PDB 1o59 chain A domain 2
Region: 112 - 122
Aligned: 11
Modelled: 11
Confidence: 7.5%
Identity: 18%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Allantoicase repeat
Phyre2
| 17 |
|
PDB 1aop chain A domain 2
Region: 111 - 144
Aligned: 34
Modelled: 34
Confidence: 7.3%
Identity: 15%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: Duplicated SiR/NiR-like domains 1 and 3
Phyre2
| 18 |
|
PDB 2q8z chain A domain 1
Region: 169 - 205
Aligned: 37
Modelled: 37
Confidence: 6.9%
Identity: 16%
Fold: TIM beta/alpha-barrel
Superfamily: Ribulose-phoshate binding barrel
Family: Decarboxylase
Phyre2
| 19 |
|
PDB 6ehr chain D
Region: 28 - 43
Aligned: 16
Modelled: 16
Confidence: 6.9%
Identity: 44%
PDB header:signaling protein
Chain: D: PDB Molecule:ragulator complex protein lamtor4;
PDBTitle: the crystal structure of the human lamtor-raga ctd-ragc ctd complex
Phyre2
| 20 |
|
PDB 1o59 chain A domain 1
Region: 111 - 122
Aligned: 12
Modelled: 12
Confidence: 5.9%
Identity: 33%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Allantoicase repeat
Phyre2
| 21 |
|
| 22 |
|