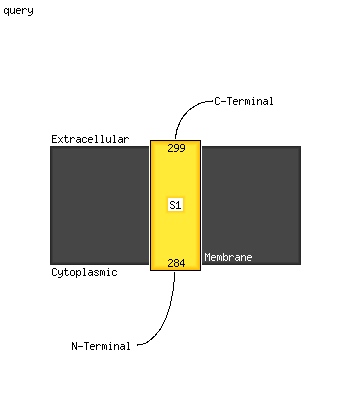
| 1 | c6aefB_
|
|
 |
100.0 |
51 |
PDB header:transferase
Chain: B: PDB Molecule:polyketide synthase associated protein papa2;
PDBTitle: papa2 acyl transferase
|
|
|
|
| 2 | c6n8eA_
|
|
 |
100.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:holo-obif1;
PDBTitle: crystal structure of holo-obif1, a five domain nonribosomal peptide2 synthetase from burkholderia diffusa
|
|
|
|
| 3 | c2vsqA_
|
|
 |
100.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:surfactin synthetase subunit 3;
PDBTitle: structure of surfactin a synthetase c (srfa-c), a nonribosomal peptide2 synthetase termination module
|
|
|
|
| 4 | c6p1jA_
|
|
 |
100.0 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:txo2;
PDBTitle: the structure of condensation and adenylation domains of teixobactin-2 producing nonribosomal peptide synthetase txo2 serine module
|
|
|
|
| 5 | c5u89A_
|
|
 |
100.0 |
15 |
PDB header:hydrolase/inhibitor
Chain: A: PDB Molecule:amino acid adenylation domain protein;
PDBTitle: crystal structure of a cross-module fragment from the dimodular nrps2 dhbf
|
|
|
|
| 6 | c4zxiA_
|
|
 |
100.0 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:tyrocidine synthetase 3;
PDBTitle: crystal structure of holo-ab3403 a four domain nonribosomal peptide2 synthetase bound to amp and glycine
|
|
|
|
| 7 | c5ja2A_
|
|
 |
100.0 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:enterobactin synthase component f;
PDBTitle: entf, a terminal nonribosomal peptide synthetase module bound to the2 non-native mbth-like protein pa2412
|
|
|
|
| 8 | c4zxjA_
|
|
 |
100.0 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:enterobactin synthase component f;
PDBTitle: crystal structure of holo-entf a nonribosomal peptide synthetase in2 the thioester-forming conformation
|
|
|
|
| 9 | c6ozvA_
|
|
 |
100.0 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:txo1;
PDBTitle: the structure of condensation and adenylation domains of teixobactin-2 producing nonribosomal peptide synthetase txo1 serine module in3 complex with amp
|
|
|
|
| 10 | c4tx3B_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide synthetase, module 7;
PDBTitle: complex of the x-domain and oxyb from teicoplanin biosynthesis
|
|
|
|
| 11 | c6m7lB_
|
|
 |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:putative non-ribosomal peptide synthetase;
PDBTitle: complex of oxya with the x-domain from gpa biosynthesis
|
|
|
|
| 12 | c5t81A_
|
|
 |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:epob;
PDBTitle: rhombohedral crystal form of the epob nrps cyclization-docking2 bidomain from sorangium cellulosum
|
|
|
|
| 13 | c2jgpA_
|
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:tyrocidine synthetase 3;
PDBTitle: structure of the tycc5-6 pcp-c bidomain of the tyrocidine synthetase2 tycc
|
|
|
|
| 14 | c6cgoB_
|
|
 |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:condensation domain protein;
PDBTitle: molecular basis for condensation domain-mediated chain release from2 the enacyloxin polyketide synthase
|
|
|
|
| 15 | c4jn3B_
|
|
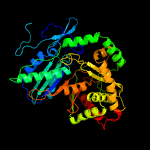 |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cda peptide synthetase i;
PDBTitle: crystal structures of the first condensation domain of the cda2 synthetase
|
|
|
|
| 16 | c2xhgA_
|
|
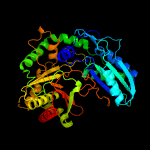 |
100.0 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:tyrocidine synthetase a;
PDBTitle: crystal structure of the epimerization domain from the initiation2 module of tyrocidine biosynthesis
|
|
|
|
| 17 | c4znmB_
|
|
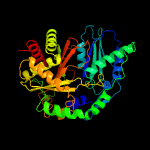 |
100.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:c-domain type ii peptide synthetase;
PDBTitle: crystal structure of sgcc5 protein from streptomyces globisporus (apo2 form)
|
|
|
|
| 18 | c5t3eA_
|
|
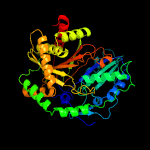 |
100.0 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:bacillamide synthetase heterocyclization domain;
PDBTitle: crystal structure of a nonribosomal peptide synthetase2 heterocyclization domain.
|
|
|
|
| 19 | c6ad3A_
|
|
 |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:lovastatin nonaketide synthase moka;
PDBTitle: structural characterization of the condensation domain from monacolin2 k polyketide synthase moka
|
|
|
|
| 20 | c5m6pB_
|
|
 |
100.0 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:tyrocidine synthase 2;
PDBTitle: crystal structure of the epimerization domain from module 3 of2 tyrocidine synthetase b, tycb3(e)
|
|
|
|
| 21 | c1l5aA_ |
|
not modelled |
100.0 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:amide synthase;
PDBTitle: crystal structure of vibh, an nrps condensation enzyme
|
|
|
| 22 | c5dijA_ |
|
not modelled |
100.0 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:tqaa;
PDBTitle: the crystal structure of ct
|
|
|
| 23 | c4hvmC_ |
|
not modelled |
100.0 |
14 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:tlmii;
PDBTitle: crystal structure of tallysomycin biosynthesis protein tlmii
|
|
|
| 24 | c3fotA_ |
|
not modelled |
100.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:15-o-acetyltransferase;
PDBTitle: structural and functional characterization of tri3 trichothecene 15-o-2 acetyltransferase from fusarium sporotrichioides
|
|
|
| 25 | c1q9jA_ |
|
not modelled |
100.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:polyketide synthase associated protein 5;
PDBTitle: structure of polyketide synthase associated protein 5 from2 mycobacterium tuberculosis
|
|
|
| 26 | c6chjB_ |
|
not modelled |
100.0 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:diacylglycerol o-acyltransferase;
PDBTitle: wax ester synthase/diacylglycerol acyltransferase from marinobacter2 aquaeolei vt8
|
|
|
| 27 | d1l5aa2 |
|
not modelled |
100.0 |
7 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:NRPS condensation domain (amide synthase) |
|
|
| 28 | d1l5aa1 |
|
not modelled |
100.0 |
12 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:NRPS condensation domain (amide synthase) |
|
|
| 29 | c6dd2A_ |
|
not modelled |
100.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:probable hydroxycinnamoyl transferase;
PDBTitle: crystal structure of selaginella moellendorffii hct
|
|
|
| 30 | d1q9ja1 |
|
not modelled |
99.9 |
17 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:NRPS condensation domain (amide synthase) |
|
|
| 31 | d1q9ja2 |
|
not modelled |
99.9 |
10 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:NRPS condensation domain (amide synthase) |
|
|
| 32 | c4g0bA_ |
|
not modelled |
99.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:hydroxycinnamoyl-coa shikimate/quinate
PDBTitle: structure of native hct from coffea canephora
|
|
|
| 33 | c2e1uA_ |
|
not modelled |
99.8 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:acyl transferase;
PDBTitle: crystal structure of dendranthema morifolium dmat
|
|
|
| 34 | c6eqoB_ |
|
not modelled |
99.8 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:acetyl-coenzyme a synthetase;
PDBTitle: tri-functional propionyl-coa synthase of erythrobacter sp. nap1 with2 bound nadp+ and phosphomethylphosphonic acid adenylate ester
|
|
|
| 35 | c5es8A_ |
|
not modelled |
99.8 |
9 |
PDB header:ligase
Chain: A: PDB Molecule:linear gramicidin synthetase subunit a;
PDBTitle: crystal structure of the initiation module of lgra in the thiolation2 state
|
|
|
| 36 | c2bghA_ |
|
not modelled |
99.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:vinorine synthase;
PDBTitle: crystal structure of vinorine synthase
|
|
|
| 37 | c2xr7A_ |
|
not modelled |
99.6 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:malonyltransferase;
PDBTitle: crystal structure of nicotiana tabacum malonyltransferase (ntmat1)2 complexed with malonyl-coa
|
|
|
| 38 | c3b2sA_ |
|
not modelled |
99.1 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:trichothecene 3-o-acetyltransferase;
PDBTitle: crystal structure of f. graminearum tri101 complexed with coenzyme a2 and deoxynivalenol
|
|
|
| 39 | c4ke4A_ |
|
not modelled |
98.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:hydroxycinnamoyl-coa:shikimate hydroxycinnamoyl
PDBTitle: elucidation of the structure and reaction mechanism of sorghum bicolor2 hydroxycinnamoyltransferase and its structural relationship to other3 coa-dependent transferases and synthases
|
|
|
| 40 | c2zbaD_ |
|
not modelled |
98.3 |
8 |
PDB header:transferase
Chain: D: PDB Molecule:trichothecene 3-o-acetyltransferase;
PDBTitle: crystal structure of f. sporotrichioides tri101 complexed with2 coenzyme a and t-2
|
|
|
| 41 | c2i9dC_ |
|
not modelled |
84.7 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: chloramphenicol acetyltransferase
|
|
|
| 42 | c3rqcB_ |
|
not modelled |
57.8 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:probable lipoamide acyltransferase;
PDBTitle: crystal structure of the catalytic core of the 2-oxoacid dehydrogenase2 multienzyme complex from thermoplasma acidophilum
|
|
|
| 43 | d1nija2 |
|
not modelled |
49.3 |
19 |
Fold:Hypothetical protein YjiA, C-terminal domain
Superfamily:Hypothetical protein YjiA, C-terminal domain
Family:Hypothetical protein YjiA, C-terminal domain |
|
|
| 44 | d1scza_ |
|
not modelled |
43.9 |
22 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 45 | c3l60A_ |
|
not modelled |
36.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:branched-chain alpha-keto acid dehydrogenase;
PDBTitle: crystal structure of branched-chain alpha-keto acid dehydrogenase2 subunit e2 from mycobacterium tuberculosis
|
|
|
| 46 | c3b8kA_ |
|
not modelled |
28.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:dihydrolipoyllysine-residue acetyltransferase;
PDBTitle: structure of the truncated human dihydrolipoyl acetyltransferase (e2)
|
|
|
| 47 | c1vs3B_ |
|
not modelled |
25.7 |
33 |
PDB header:isomerase
Chain: B: PDB Molecule:trna pseudouridine synthase a;
PDBTitle: crystal structure of the trna pseudouridine synthase trua from thermus2 thermophilus hb8
|
|
|
| 48 | c4n72B_ |
|
not modelled |
24.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate dehydrogenase (dihydrolipoyltransacetylase
PDBTitle: catalytic domain from dihydrolipoamide acetyltransferase of pyruvate2 dehydrogenase from escherichia coli
|
|
|
| 49 | d1dpba_ |
|
not modelled |
22.2 |
9 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 50 | d1q23a_ |
|
not modelled |
21.9 |
10 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 51 | d1b5sa_ |
|
not modelled |
20.6 |
13 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 52 | d1dj0a_ |
|
not modelled |
14.1 |
17 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase I TruA |
|
|
| 53 | c5hypB_ |
|
not modelled |
13.6 |
22 |
PDB header:immune system
Chain: B: PDB Molecule:m28 protein;
PDBTitle: structure of human c4b-binding protein alpha cain ccp domains 1 and 22 in complex with the hypervariable region of group a streptococcus m283 protein
|
|
|
| 54 | c5wmmA_ |
|
not modelled |
13.0 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:nrps;
PDBTitle: crystal structure of an adenylation domain interrupted by a2 methylation domain (ama4) from nonribosomal peptide synthetase tios
|
|
|
| 55 | c2ii4C_ |
|
not modelled |
11.4 |
25 |
PDB header:transferase
Chain: C: PDB Molecule:lipoamide acyltransferase component of branched-chain
PDBTitle: crystal structure of a cubic core of the dihydrolipoamide2 acyltransferase (e2b) component in the branched-chain alpha-ketoacid3 dehydrogenase complex (bckdc), coenzyme a-bound form
|
|
|
| 56 | c4gr5B_ |
|
not modelled |
9.4 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:non-ribosomal peptide synthetase;
PDBTitle: crystal structure of slgn1deltaasub in complex with ampcpp
|
|
|
| 57 | c1nijA_ |
|
not modelled |
8.4 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yjia;
PDBTitle: yjia protein
|
|
|
| 58 | d1g4ma1 |
|
not modelled |
8.1 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Arrestin/Vps26-like |
|
|
| 59 | d2h1ta1 |
|
not modelled |
8.0 |
33 |
Fold:Spiral beta-roll
Superfamily:PA1994-like
Family:PA1994-like |
|
|
| 60 | c3maeA_ |
|
not modelled |
7.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-oxoisovalerate dehydrogenase e2 component,
PDBTitle: crystal structure of probable dihydrolipoamide acetyltransferase from2 listeria monocytogenes 4b f2365
|
|
|
| 61 | c2k5jB_ |
|
not modelled |
7.2 |
3 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein yiif;
PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
|
|
|
| 62 | c2p0yA_ |
|
not modelled |
7.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus plantarum.2 northeast structural genomics consortium target lpr6
|
|
|
| 63 | c1o7dC_ |
|
not modelled |
6.8 |
28 |
PDB header:hydrolase
Chain: C: PDB Molecule:lysosomal alpha-mannosidase;
PDBTitle: the structure of the bovine lysosomal a-mannosidase suggests a novel2 mechanism for low ph activation
|
|
|
| 64 | d1zh2a1 |
|
not modelled |
6.1 |
19 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 65 | d3claa_ |
|
not modelled |
5.7 |
15 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 66 | d1p94a_ |
|
not modelled |
5.6 |
20 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
|
|
| 67 | c6bk9A_ |
|
not modelled |
5.5 |
8 |
PDB header:signaling protein
Chain: A: PDB Molecule:visual arrestin;
PDBTitle: crystal structure of squid arrestin
|
|
|
| 68 | d1u8sa2 |
|
not modelled |
5.4 |
19 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
|
|