| 1 |
|
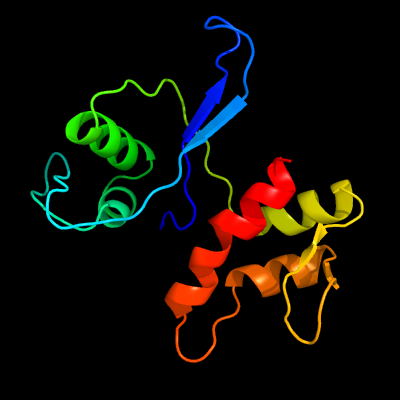
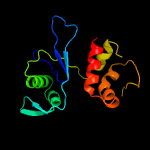
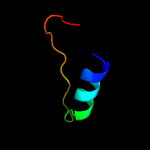
PDB 3pe5 chain B
Region: 6 - 134
Aligned: 128
Modelled: 129
Confidence: 100.0%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: three-dimensional structure of protein a7vv38_9clot from clostridium2 leptum dsm 753, northeast structural genomics consortium target3 qlr103
Phyre2
| 2 |
|
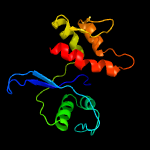
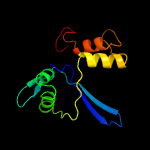
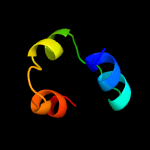
PDB 4de8 chain A
Region: 3 - 134
Aligned: 130
Modelled: 132
Confidence: 100.0%
Identity: 26%
PDB header:membrane protein
Chain: A: PDB Molecule:cps2a;
PDBTitle: lytr-cps2a-psr family protein with bound octaprenyl monophosphate2 lipid
Phyre2
| 3 |
|
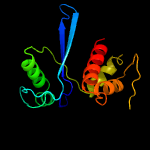
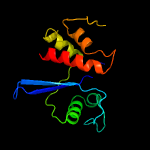
PDB 3nro chain A
Region: 7 - 134
Aligned: 127
Modelled: 128
Confidence: 100.0%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo1026 protein;
PDBTitle: crystal structure of putative transcriptional factor lmo1026 from2 listeria monocytogenes (fragment 52-321), northeast structural3 genomics consortium target lmr194
Phyre2
| 4 |
|
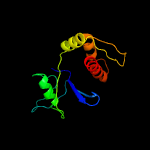
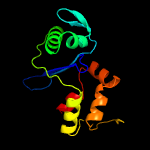
PDB 3okz chain B
Region: 3 - 134
Aligned: 131
Modelled: 132
Confidence: 100.0%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein gbs0355;
PDBTitle: crystal structure of protein gbs0355 from streptococcus agalactiae,2 northeast structural genomics consortium target sar127
Phyre2
| 5 |
|
PDB 5v8c chain A
Region: 6 - 123
Aligned: 114
Modelled: 118
Confidence: 100.0%
Identity: 25%
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: lytr-csp2a-psr enzyme from actinomyces oris
Phyre2
| 6 |
|
PDB 3qfi chain A
Region: 7 - 134
Aligned: 127
Modelled: 128
Confidence: 100.0%
Identity: 29%
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: x-ray crystal structure of transcriptional regulator (ef0465) from2 enterococcus faecalis, northeast structural genomics consortium3 target efr190
Phyre2
| 7 |
|
PDB 3mej chain A
Region: 6 - 134
Aligned: 125
Modelled: 129
Confidence: 100.0%
Identity: 38%
PDB header:transcriptional regulator
Chain: A: PDB Molecule:transcriptional regulator ywtf;
PDBTitle: crystal structure of putative transcriptional regulator ywtf from2 bacillus subtilis, northeast structural genomics consortium target3 sr736
Phyre2
| 8 |
|
PDB 3owq chain B
Region: 6 - 134
Aligned: 125
Modelled: 129
Confidence: 100.0%
Identity: 32%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lin1025 protein;
PDBTitle: x-ray structure of lin1025 protein from listeria innocua, northeast2 structural genomics consortium target lkr164
Phyre2
| 9 |
|
PDB 4obm chain A
Region: 6 - 134
Aligned: 124
Modelled: 124
Confidence: 100.0%
Identity: 13%
PDB header:transcription regulator
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative transcription regulator (eubsir_01389)2 from eubacterium siraeum dsm 15702 at 2.15 a resolution
Phyre2
| 10 |
|
PDB 3nxh chain A
Region: 6 - 134
Aligned: 115
Modelled: 129
Confidence: 100.0%
Identity: 27%
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator yvhj;
PDBTitle: crystal structure of the transcriptional regulator yvhj from bacillus2 subtilis. northeast structural genomics consortium target sr735.
Phyre2
| 11 |
|
PDB 4qn9 chain A
Region: 77 - 112
Aligned: 36
Modelled: 36
Confidence: 12.0%
Identity: 22%
PDB header:hydrolase
Chain: A: PDB Molecule:n-acyl-phosphatidylethanolamine-hydrolyzing phospholipase
PDBTitle: structure of human nape-pld
Phyre2
| 12 |
|
PDB 2jui chain A
Region: 78 - 93
Aligned: 16
Modelled: 16
Confidence: 9.7%
Identity: 31%
PDB header:toxin
Chain: A: PDB Molecule:plne;
PDBTitle: three-dimensional structure of the two peptides that2 constitute the two-peptide bacteriocin plantaracin ef
Phyre2
| 13 |
|
PDB 2pju chain D
Region: 47 - 87
Aligned: 41
Modelled: 41
Confidence: 7.9%
Identity: 7%
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon regulatory protein2 prpr
Phyre2
| 14 |
|
PDB 3zpp chain A
Region: 76 - 104
Aligned: 29
Modelled: 29
Confidence: 7.7%
Identity: 28%
PDB header:cell adhesion
Chain: A: PDB Molecule:cell wall surface anchor family protein;
PDBTitle: structure of the streptococcus pneumoniae surface protein2 and adhesin pfba
Phyre2
| 15 |
|
PDB 1vqo chain N domain 1
Region: 77 - 127
Aligned: 46
Modelled: 51
Confidence: 6.2%
Identity: 33%
Fold: Ribonuclease H-like motif
Superfamily: Translational machinery components
Family: Ribosomal protein L18 and S11
Phyre2
| 16 |
|
PDB 2pju chain A domain 1
Region: 47 - 87
Aligned: 41
Modelled: 41
Confidence: 6.2%
Identity: 7%
Fold: Chelatase-like
Superfamily: PrpR receptor domain-like
Family: PrpR receptor domain-like
Phyre2
| 17 |
|
PDB 2r8r chain B
Region: 59 - 97
Aligned: 39
Modelled: 39
Confidence: 5.3%
Identity: 21%
PDB header:transferase
Chain: B: PDB Molecule:sensor protein;
PDBTitle: crystal structure of the n-terminal region (19..243) of sensor protein2 kdpd from pseudomonas syringae pv. tomato str. dc3000
Phyre2
| 18 |
|
PDB 1bhe chain A
Region: 76 - 102
Aligned: 27
Modelled: 27
Confidence: 5.1%
Identity: 22%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pectin lyase-like
Family: Galacturonase
Phyre2
| 19 |
|
PDB 3ejb chain C
Region: 56 - 87
Aligned: 32
Modelled: 32
Confidence: 5.0%
Identity: 9%
PDB header:oxidoreductase/lipid transport
Chain: C: PDB Molecule:acyl carrier protein;
PDBTitle: crystal structure of p450bioi in complex with tetradecanoic acid2 ligated acyl carrier protein
Phyre2