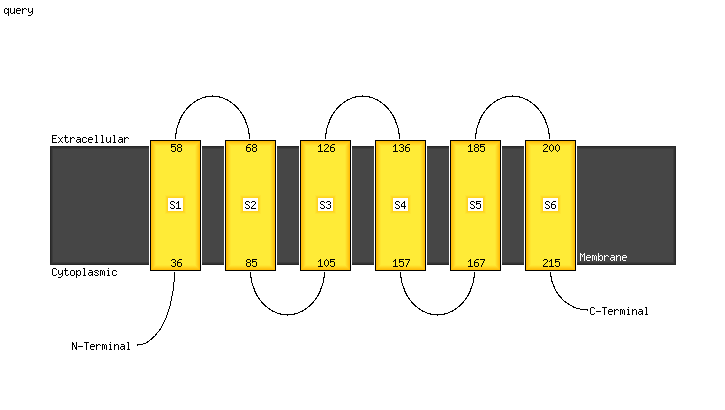
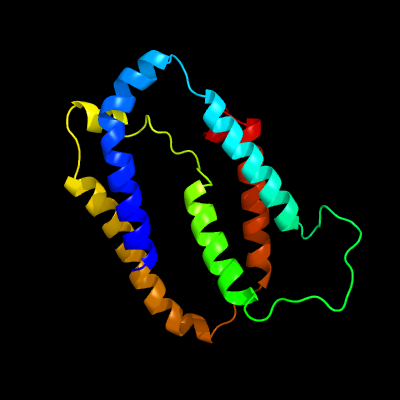
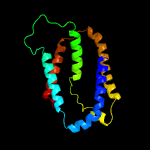
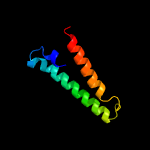
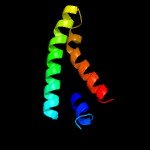
1 c5vkvA_
95.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccda;PDBTitle: solution nmr structure of the membrane electron transporter ccda
2 c6mjpF_
63.8
11
PDB header: lipid transportChain: F: PDB Molecule: fig000988: predicted permease;PDBTitle: lptb(e163q)fgc from vibrio cholerae
3 c5x5yG_
56.6
17
PDB header: membrane proteinChain: G: PDB Molecule: uncharacterized protein;PDBTitle: a membrane protein complex
4 c5x5yF_
53.0
10
PDB header: membrane proteinChain: F: PDB Molecule: uncharacterized protein;PDBTitle: a membrane protein complex
5 c5l75G_
44.3
14
PDB header: transport proteinChain: G: PDB Molecule: fig000906: predicted permease;PDBTitle: a protein structure
6 c3hd6A_
44.2
20
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: ammonium transporter rh type c;PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
7 c6mjpG_
44.0
16
PDB header: lipid transportChain: G: PDB Molecule: lps export abc transporter permease lptg;PDBTitle: lptb(e163q)fgc from vibrio cholerae
8 c5ze3B_
36.2
40
PDB header: oxidoreductaseChain: B: PDB Molecule: lysyl oxidase homolog 2;PDBTitle: crystal structure of human lysyl oxidase-like 2 (hloxl2) in a2 precursor state
9 c5l75F_
35.9
6
PDB header: transport proteinChain: F: PDB Molecule: fig000988: predicted permease;PDBTitle: a protein structure
10 c5aezA_
31.1
15
PDB header: membrane proteinChain: A: PDB Molecule: mep2;PDBTitle: crystal structure of candida albicans mep2
11 c5a43B_
30.3
9
PDB header: transport proteinChain: B: PDB Molecule: putative fluoride ion transporter crcb;PDBTitle: crystal structure of a dual topology fluoride ion channel.
12 d1mxaa1
30.0
55
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase
13 d2p02a1
28.5
55
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase
14 d1qm4a1
28.3
55
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase
15 d1ei5a2
28.1
62
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains
16 d2e74f1
27.9
57
Fold: Single transmembrane helixSuperfamily: PetM subunit of the cytochrome b6f complexFamily: PetM subunit of the cytochrome b6f complex
17 c4mbyB_
23.5
24
PDB header: viral proteinChain: B: PDB Molecule: major capsid protein vp1;PDBTitle: structure of b-lymphotropic polyomavirus vp1 in complex with 3'-2 sialyllactose
18 c5aexJ_
21.3
13
PDB header: membrane proteinChain: J: PDB Molecule: ammonium transporter mep2;PDBTitle: crystal structure of saccharomyces cerevisiae mep2
19 c5vn4A_
19.5
42
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase, putative;PDBTitle: crystal structure of adenine phosphoribosyl transferase from2 trypanosoma brucei in complex with amp, pyrophosphate, and ribose-5-3 phosphate
20 c3c1iA_
18.1
25
PDB header: transport proteinChain: A: PDB Molecule: ammonia channel;PDBTitle: substrate binding, deprotonation and selectivity at the periplasmic2 entrance of the e. coli ammonia channel amtb
21 c2kr6A_
not modelled
16.5
13
PDB header: hydrolaseChain: A: PDB Molecule: presenilin-1;PDBTitle: solution structure of presenilin-1 ctf subunit
22 c2obvA_
not modelled
15.7
55
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine synthetase isoform type-1;PDBTitle: crystal structure of the human s-adenosylmethionine synthetase 1 in2 complex with the product
23 c3imlB_
not modelled
15.6
45
PDB header: transferaseChain: B: PDB Molecule: s-adenosylmethionine synthetase;PDBTitle: crystal structure of s-adenosylmethionine synthetase from burkholderia2 pseudomallei
24 c3rv2B_
not modelled
15.5
55
PDB header: transferaseChain: B: PDB Molecule: s-adenosylmethionine synthase;PDBTitle: crystal structure of s-adenosylmethionine synthetase from2 mycobacterium marinum
25 c5h9uC_
not modelled
15.5
45
PDB header: transferaseChain: C: PDB Molecule: s-adenosylmethionine synthase;PDBTitle: crystal structure of a thermostable methionine adenosyltransferase
26 c1rg9D_
not modelled
15.5
55
PDB header: transferaseChain: D: PDB Molecule: s-adenosylmethionine synthetase;PDBTitle: s-adenosylmethionine synthetase complexed with sam and ppnp
27 c4odjA_
not modelled
15.0
45
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine synthase;PDBTitle: crystal structure of a putative s-adenosylmethionine synthetase from2 cryptosporidium hominis in complex with s-adenosyl-methionine
28 c6hwhX_
not modelled
14.9
23
PDB header: electron transportChain: X: PDB Molecule: cytochrome c oxidase polypeptide 4;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
29 c3so4C_
not modelled
14.6
45
PDB header: transferaseChain: C: PDB Molecule: methionine-adenosyltransferase;PDBTitle: methionine-adenosyltransferase from entamoeba histolytica
30 c4le5A_
not modelled
14.5
36
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine synthetase;PDBTitle: structure of an unusual s-adenosylmethionine synthetase from2 campylobacter jejuni
31 c4k1cB_
not modelled
13.6
13
PDB header: membrane protein/metal transportChain: B: PDB Molecule: vacuolar calcium ion transporter;PDBTitle: vcx1 calcium/proton exchanger
32 d1k8ib2
not modelled
13.2
64
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain
33 c4k1cA_
not modelled
13.1
13
PDB header: membrane protein/metal transportChain: A: PDB Molecule: vacuolar calcium ion transporter;PDBTitle: vcx1 calcium/proton exchanger
34 c3j1zP_
not modelled
12.7
13
PDB header: metal transportChain: P: PDB Molecule: cation efflux family protein;PDBTitle: inward-facing conformation of the zinc transporter yiip revealed by2 cryo-electron microscopy
35 d1u7ga_
not modelled
12.6
19
Fold: Ammonium transporterSuperfamily: Ammonium transporterFamily: Ammonium transporter
36 c2l9uB_
not modelled
11.7
12
PDB header: membrane proteinChain: B: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
37 c2l9uA_
not modelled
11.7
12
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
38 c2lx0A_
not modelled
11.1
29
PDB header: membrane proteinChain: A: PDB Molecule: membrane fusion protein p14;PDBTitle: arced helix (arch) nmr structure of the reovirus p14 fusion-associated2 small transmembrane (fast) protein transmembrane domain (tmd) in3 dodecyl phosphocholine (dpc) micelles
39 c2zt9F_
not modelled
10.9
50
PDB header: photosynthesisChain: F: PDB Molecule: cytochrome b6-f complex subunit 7;PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
40 c4h44F_
not modelled
10.9
50
PDB header: photosynthesisChain: F: PDB Molecule: cytochrome b6-f complex subunit 7;PDBTitle: 2.70 a cytochrome b6f complex structure from nostoc pcc 7120
41 c4ogqF_
not modelled
10.9
50
PDB header: electron transportChain: F: PDB Molecule: cytochrome b6-f complex subunit 7;PDBTitle: internal lipid architecture of the hetero-oligomeric cytochrome b6f2 complex
42 c4iu8A_
not modelled
10.8
9
PDB header: transport proteinChain: A: PDB Molecule: nitrite extrusion protein 2;PDBTitle: crystal structure of a membrane transporter (selenomethionine2 derivative)
43 c2ks1A_
not modelled
10.4
25
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
44 c2jwaA_
not modelled
10.4
25
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
45 c5aj3m_
not modelled
9.9
50
PDB header: ribosomeChain: M: PDB Molecule: PDBTitle: structure of the small subunit of the mammalian mitoribosome
46 d1yy9a4
not modelled
9.4
67
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Growth factor receptor domainFamily: Growth factor receptor domain
47 c3uc8A_
not modelled
9.3
86
PDB header: de novo proteinChain: A: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - tetragonal crystal form
48 c3uc8B_
not modelled
9.3
86
PDB header: de novo proteinChain: B: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - tetragonal crystal form
49 c3uc7E_
not modelled
9.3
86
PDB header: de novo proteinChain: E: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - monoclinic crystal form
50 c2ll5A_
not modelled
9.3
86
PDB header: de novo proteinChain: A: PDB Molecule: cyclo-tc1;PDBTitle: cyclo-tc1 trp-cage
51 c3uc7A_
not modelled
9.3
86
PDB header: de novo proteinChain: A: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - monoclinic crystal form
52 c3uc7D_
not modelled
9.3
86
PDB header: de novo proteinChain: D: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - monoclinic crystal form
53 c3uc7C_
not modelled
9.3
86
PDB header: de novo proteinChain: C: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - monoclinic crystal form
54 c3uc7B_
not modelled
9.3
86
PDB header: de novo proteinChain: B: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - monoclinic crystal form
55 c3uc7F_
not modelled
9.3
86
PDB header: de novo proteinChain: F: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - monoclinic crystal form
56 c3uc8C_
not modelled
9.3
86
PDB header: de novo proteinChain: C: PDB Molecule: cyclo-tc1;PDBTitle: trp-cage cyclo-tc1 - tetragonal crystal form
57 c6nbxG_
not modelled
8.7
12
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: t.elongatus ndh (data-set 2)
58 c3wl7A_
not modelled
8.4
70
PDB header: hydrolaseChain: A: PDB Molecule: oxidized polyvinyl alcohol hydrolase;PDBTitle: the complex structure of poph s172c with ligand, aca
59 d2de6a2
not modelled
8.1
40
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Ring hydroxylating alpha subunit catalytic domain
60 c4g5sZ_
not modelled
7.9
50
PDB header: cell cycle/signaling proteinChain: Z: PDB Molecule: g-protein-signaling modulator 2;PDBTitle: structure of lgn gl3/galphai3 complex
61 c5twaD_
not modelled
7.8
71
PDB header: apoptosisChain: D: PDB Molecule: bak-2 protein;PDBTitle: crystal structure of geodia cydonium bhp2 in complex with lubomirskia2 baicalensis bak-2
62 c5twaC_
not modelled
7.8
71
PDB header: apoptosisChain: C: PDB Molecule: bak-2 protein;PDBTitle: crystal structure of geodia cydonium bhp2 in complex with lubomirskia2 baicalensis bak-2
63 d1fftb2
not modelled
7.8
14
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region
64 c2qz5A_
not modelled
7.6
33
PDB header: signaling protein, lipid binding proteinChain: A: PDB Molecule: axin interactor, dorsalization associatedPDBTitle: crystal structure of the c-terminal domain of aida
65 d1vpsa_
not modelled
7.3
21
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group I dsDNA virusesFamily: Papovaviridae-like VP
66 c4fmiN_
not modelled
7.1
18
PDB header: viral proteinChain: N: PDB Molecule: vp1;PDBTitle: merkel cell polyomavirus vp1 in complex with 3'-sialyllactosamine
67 c4g5sG_
not modelled
6.7
55
PDB header: cell cycle/signaling proteinChain: G: PDB Molecule: g-protein-signaling modulator 2;PDBTitle: structure of lgn gl3/galphai3 complex
68 c3a5pD_
not modelled
6.6
20
PDB header: sugar binding proteinChain: D: PDB Molecule: haemagglutinin i;PDBTitle: crystal structure of hemagglutinin
69 c2czsB_
not modelled
6.5
23
PDB header: electron transportChain: B: PDB Molecule: cytochrome c, putative;PDBTitle: crystal structure analysis of the diheme c-type cytochrome dhc2
70 c4s1kA_
not modelled
6.4
38
PDB header: viral proteinChain: A: PDB Molecule: polyhedrin;PDBTitle: structure of uranotaenia sapphirina cypovirus (cpv17) polyhedrin at2 100 k
71 c3j39Q_
not modelled
6.3
31
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l18;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
72 d1prtf_
not modelled
6.2
38
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits
73 c2ka1B_
not modelled
6.2
42
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
74 c2ka2A_
not modelled
6.2
42
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
75 d2bida_
not modelled
6.1
57
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death
76 c5tcxA_
not modelled
6.1
20
PDB header: cell invasionChain: A: PDB Molecule: cd81 antigen;PDBTitle: crystal structure of human tetraspanin cd81
77 c6g72A_
not modelled
5.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-ubiquinone oxidoreductase chain 3;PDBTitle: mouse mitochondrial complex i in the deactive state
78 c2n90A_
not modelled
5.8
22
PDB header: transferaseChain: A: PDB Molecule: high affinity nerve growth factor receptor;PDBTitle: trka transmembrane domain nmr structure in dpc micelles
79 c2n90B_
not modelled
5.8
22
PDB header: transferaseChain: B: PDB Molecule: high affinity nerve growth factor receptor;PDBTitle: trka transmembrane domain nmr structure in dpc micelles
80 c1bzkA_
not modelled
5.7
25
PDB header: transport proteinChain: A: PDB Molecule: protein (band 3 anion transport protein);PDBTitle: structural studies on the effects of the deletion in the2 red cell anion exchanger (band3, ae1) associated with3 south east asian ovalocytosis.
81 c2n4xA_
not modelled
5.7
16
PDB header: membrane proteinChain: A: PDB Molecule: cytochrome c-type biogenesis protein (ccda);PDBTitle: structure of the transmembrane electron transporter ccda
82 c6qvcB_
not modelled
5.6
8
PDB header: membrane proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: cryoem structure of the human clc-1 chloride channel, cbs state 1
83 d1ddba_
not modelled
5.3
57
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death
84 c2lonA_
not modelled
5.2
12
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1b;PDBTitle: backbone structure of human membrane protein higd1b
85 c5aexB_
not modelled
5.2
11
PDB header: membrane proteinChain: B: PDB Molecule: ammonium transporter mep2;PDBTitle: crystal structure of saccharomyces cerevisiae mep2
86 d1vd4a_
not modelled
5.2
33
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain
87 c6iedA_
not modelled
5.2
6
PDB header: membrane proteinChain: A: PDB Molecule: heme a synthase;PDBTitle: crystal structure of heme a synthase from bacillus subtilis