| 1 | c1nxjA_
|
|
 |
100.0 |
100 |
PDB header:unknown function
Chain: A: PDB Molecule:probable s-adenosylmethionine:2-
PDBTitle: structure of rv3853 from mycobacterium tuberculosis
|
|
|
|
| 2 | d1nxja_
|
|
 |
100.0 |
100 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
|
| 3 | c2pcnA_
|
|
 |
100.0 |
44 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine:2-demethylmenaquinone
PDBTitle: crystal structure of s-adenosylmethionine: 2-dimethylmenaquinone2 methyltransferase (gk_1813) from geobacillus kaustophilus hta426
|
|
|
|
| 4 | d1vi4a_
|
|
 |
100.0 |
47 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
|
| 5 | d1q5xa_
|
|
 |
100.0 |
45 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
|
| 6 | c3c8oB_
|
|
 |
100.0 |
44 |
PDB header:hydrolase regulator
Chain: B: PDB Molecule:regulator of ribonuclease activity a;
PDBTitle: the crystal structure of rraa from pao1
|
|
|
|
| 7 | c3nojA_
|
|
 |
100.0 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate
PDBTitle: the structure of hmg/cha aldolase from the protocatechuate degradation2 pathway of pseudomonas putida
|
|
|
|
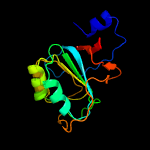
| 8 | d1j3la_
|
|
 |
100.0 |
45 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
|
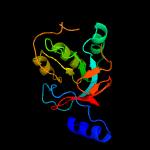
| 9 | c5ir2A_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellulase;
PDBTitle: crystal structure of novel cellulases from microbes associated with2 the gut ecosystem
|
|
|
|
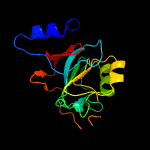
| 10 | c3k4iC_
|
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
|
|
|
|
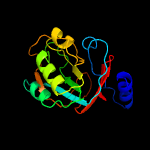
| 11 | c5x15C_
|
|
 |
100.0 |
28 |
PDB header:transferase inhibitor
Chain: C: PDB Molecule:putative transferase;
PDBTitle: crystal structure of streptomyces coelicolor rraas2, an unusual member2 of the rnase es inhibitor rraa protein family
|
|
|
|
| 12 | c2c5qE_
|
|
 |
100.0 |
20 |
PDB header:structural genomics,unknown function
Chain: E: PDB Molecule:rraa-like protein yer010c;
PDBTitle: crystal structure of yeast yer010cp
|
|
|
|
| 13 | d2gp4a1
|
|
 |
72.3 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:IlvD/EDD C-terminal domain-like |
|
|
|
| 14 | c2gp4A_
|
|
 |
70.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
|
|
|
| 15 | c2gp4B_
|
|
 |
66.8 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
|
|
|
| 16 | c6ovtD_
|
|
 |
56.9 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:dihydroxy-acid dehydratase;
PDBTitle: crystal structure of ilvd from mycobacterium tuberculosis
|
|
|
|
| 17 | c5j84A_
|
|
 |
55.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase;
PDBTitle: crystal structure of l-arabinonate dehydratase in holo-form
|
|
|
|
| 18 | c5oynB_
|
|
 |
53.1 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:dehydratase, ilvd/edd family;
PDBTitle: crystal structure of d-xylonate dehydratase in holo-form
|
|
|
|
| 19 | d1kbla2
|
|
 |
48.9 |
23 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
|
| 20 | c5ze4A_
|
|
 |
48.4 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
|
|
|
|
| 21 | c5hdjA_ |
|
not modelled |
47.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nfra1;
PDBTitle: structure of b. megaterium nfra1
|
|
|
| 22 | c5ym0A_ |
|
not modelled |
45.8 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the crystal structure of dhad
|
|
|
| 23 | d1f5va_ |
|
not modelled |
44.4 |
21 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
|
|
| 24 | c3n2sD_ |
|
not modelled |
41.6 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nadph-dependent nitro/flavin reductase;
PDBTitle: structure of nfra1 nitroreductase from b. subtilis
|
|
|
| 25 | d1e8ca2 |
|
not modelled |
41.2 |
20 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
| 26 | d1zcha1 |
|
not modelled |
39.2 |
10 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
|
|
| 27 | d1vfga2 |
|
not modelled |
32.7 |
35 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly A polymerase head domain-like |
|
|
| 28 | c5heiE_ |
|
not modelled |
32.7 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nfra2;
PDBTitle: structure of b. megaterium nfra2
|
|
|
| 29 | c3eofB_ |
|
not modelled |
32.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_213212.1) from2 bacteroides fragilis nctc 9343 at 1.99 a resolution
|
|
|
| 30 | d1bkja_ |
|
not modelled |
32.3 |
18 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
|
|
| 31 | c1vfgB_ |
|
not modelled |
31.7 |
35 |
PDB header:transferase/rna
Chain: B: PDB Molecule:poly a polymerase;
PDBTitle: crystal structure of trna nucleotidyltransferase complexed2 with a primer trna and an incoming atp analog
|
|
|
| 32 | c1ou5A_ |
|
not modelled |
30.8 |
36 |
PDB header:translation, transferase
Chain: A: PDB Molecule:trna cca-adding enzyme;
PDBTitle: crystal structure of human cca-adding enzyme
|
|
|
| 33 | c1dbgA_ |
|
not modelled |
27.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:chondroitinase b;
PDBTitle: crystal structure of chondroitinase b
|
|
|
| 34 | d1ofla_ |
|
not modelled |
26.9 |
26 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:Chondroitinase B |
|
|
| 35 | d1vbga2 |
|
not modelled |
26.9 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
| 36 | c2vpiA_ |
|
not modelled |
25.8 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase;
PDBTitle: human gmp synthetase - glutaminase domain
|
|
|
| 37 | d1ou5a2 |
|
not modelled |
25.7 |
36 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly A polymerase head domain-like |
|
|
| 38 | d1miwa2 |
|
not modelled |
19.3 |
36 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly A polymerase head domain-like |
|
|
| 39 | c6odm5_ |
|
not modelled |
17.3 |
33 |
PDB header:viral protein
Chain: 5: PDB Molecule:triplex capsid protein 1;
PDBTitle: herpes simplex virus type 1 (hsv-1) portal vertex-adjacent2 capsid/catc, asymmetric unit
|
|
|
| 40 | c4x4wB_ |
|
not modelled |
15.3 |
36 |
PDB header:rna binding protein
Chain: B: PDB Molecule:cca trna nucleotidyltransferase 1, mitochondrial;
PDBTitle: crystal structure of the full-length human mitochondrial cca-adding2 enzyme
|
|
|
| 41 | c3aqnA_ |
|
not modelled |
15.1 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) polymerase;
PDBTitle: complex structure of bacterial protein (apo form ii)
|
|
|
| 42 | c5gkdA_ |
|
not modelled |
14.9 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:alygc;
PDBTitle: structure of pl6 family alginate lyase alygc
|
|
|
| 43 | c1e8cB_ |
|
not modelled |
14.1 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-diaminopimelate
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide synthetase from2 e. coli
|
|
|
| 44 | c3mvnA_ |
|
not modelled |
13.9 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamyl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
|
|
| 45 | d1vzwa1 |
|
not modelled |
13.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 46 | c3h37B_ |
|
not modelled |
12.9 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:trna nucleotidyl transferase-related protein;
PDBTitle: the structure of cca-adding enzyme apo form i
|
|
|
| 47 | c3l7oB_ |
|
not modelled |
12.6 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
|
|
| 48 | c3tbiB_ |
|
not modelled |
11.1 |
33 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of t4 gp33 bound to e. coli rnap beta-flap domain
|
|
|
| 49 | c6q52A_ |
|
not modelled |
10.8 |
36 |
PDB header:rna binding protein
Chain: A: PDB Molecule:cca-adding enzyme;
PDBTitle: structure of a psychrophilic cca-adding enzyme in complex with cmpcpp2 at room temperature in chipx microfluidic device
|
|
|
| 50 | c3bm2B_ |
|
not modelled |
10.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:protein ydja;
PDBTitle: crystal structure of a minimal nitroreductase ydja from escherichia2 coli k12 with and without fmn cofactor
|
|
|
| 51 | c2pjmA_ |
|
not modelled |
10.4 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
|
|
| 52 | d1thfd_ |
|
not modelled |
10.2 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 53 | c3k6hB_ |
|
not modelled |
10.2 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitroreductase family protein;
PDBTitle: crystal structure of a nitroreductase family protein from2 agrobacterium tumefaciens str. c58
|
|
|
| 54 | c2xrfA_ |
|
not modelled |
9.5 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:uridine phosphorylase 2;
PDBTitle: crystal structure of human uridine phosphorylase 2
|
|
|
| 55 | c1y43B_ |
|
not modelled |
9.3 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:aspergillopepsin ii heavy chain;
PDBTitle: crystal structure of aspergilloglutamic peptidase from aspergillus2 niger
|
|
|
| 56 | c3i28A_ |
|
not modelled |
8.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:epoxide hydrolase 2;
PDBTitle: crystal structure of soluble epoxide hydrolase
|
|
|
| 57 | d1i1qb_ |
|
not modelled |
8.6 |
25 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
|
|
| 58 | c1miyB_ |
|
not modelled |
8.5 |
36 |
PDB header:translation, transferase
Chain: B: PDB Molecule:trna cca-adding enzyme;
PDBTitle: crystal structure of bacillus stearothermophilus cca-adding enzyme in2 complex with ctp
|
|
|
| 59 | d1ka9f_ |
|
not modelled |
8.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 60 | c5z9tB_ |
|
not modelled |
8.2 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:alginate lyase alyf-ou02;
PDBTitle: a new pl6 alginate lyase complex with trisaccharide
|
|
|
| 61 | c2wzvB_ |
|
not modelled |
7.6 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nfnb protein;
PDBTitle: crystal structure of the fmn-dependent nitroreductase nfnb2 from mycobacterium smegmatis
|
|
|
| 62 | d3bi1a2 |
|
not modelled |
7.5 |
16 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:PA domain
Family:PA domain |
|
|
| 63 | c2xjaD_ |
|
not modelled |
7.4 |
20 |
PDB header:ligase
Chain: D: PDB Molecule:udp-n-acetylmuramoyl-l-alanyl-d-glutamate--2,6-
PDBTitle: structure of mure from m.tuberculosis with dipeptide and adp
|
|
|
| 64 | c3cioA_ |
|
not modelled |
7.4 |
22 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:tyrosine-protein kinase etk;
PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
|
|
|
| 65 | c2lktA_ |
|
not modelled |
6.9 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:retinoic acid receptor responder protein 3;
PDBTitle: solution structure of n-terminal domain of human tig3 in 2 m urea
|
|
|
| 66 | c1vdzA_ |
|
not modelled |
6.9 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:a-type atpase subunit a;
PDBTitle: crystal structure of a-type atpase catalytic subunit a from2 pyrococcus horikoshii ot3
|
|
|
| 67 | c4gmkB_ |
|
not modelled |
6.9 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose 5-phosphate isomerase from the probiotic2 bacterium lactobacillus salivarius ucc118
|
|
|
| 68 | d1j5ya1 |
|
not modelled |
6.8 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Biotin repressor-like |
|
|
| 69 | c6cslA_ |
|
not modelled |
6.6 |
30 |
PDB header:metal binding protein
Chain: A: PDB Molecule:histidine triad protein d;
PDBTitle: pneumococcal phtd protein 269-339 fragment with bound zn(ii)
|
|
|
| 70 | c1gqqA_ |
|
not modelled |
6.5 |
22 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
|
|
|
| 71 | d2cs7a1 |
|
not modelled |
6.5 |
35 |
Fold:IL8-like
Superfamily:PhtA domain-like
Family:PhtA domain-like |
|
|
| 72 | c3mogA_ |
|
not modelled |
6.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxybutyryl-coa dehydrogenase;
PDBTitle: crystal structure of 3-hydroxybutyryl-coa dehydrogenase from2 escherichia coli k12 substr. mg1655
|
|
|
| 73 | d1bvp12 |
|
not modelled |
6.1 |
30 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Top domain of virus capsid protein |
|
|
| 74 | d1ahsa_ |
|
not modelled |
6.0 |
22 |
Fold:Viral protein domain
Superfamily:Viral protein domain
Family:Top domain of virus capsid protein |
|
|
| 75 | c6iunB_ |
|
not modelled |
5.9 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-coa hydratase/delta(3)-cis-delta(2)-trans-enoyl-coa
PDBTitle: crystal structure of enoyl-coa hydratase (ech) from ralstonia eutropha2 h16 in complex with nad
|
|
|
| 76 | c2kytA_ |
|
not modelled |
5.9 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:group xvi phospholipase a2;
PDBTitle: solution structure of the h-rev107 n-terminal domain
|
|
|
| 77 | c2d3tB_ |
|
not modelled |
5.7 |
12 |
PDB header:lyase, oxidoreductase/transferase
Chain: B: PDB Molecule:fatty oxidation complex alpha subunit;
PDBTitle: fatty acid beta-oxidation multienzyme complex from2 pseudomonas fragi, form v
|
|
|
| 78 | d1eara1 |
|
not modelled |
5.6 |
35 |
Fold:Urease metallochaperone UreE, N-terminal domain
Superfamily:Urease metallochaperone UreE, N-terminal domain
Family:Urease metallochaperone UreE, N-terminal domain |
|
|
| 79 | c3bemA_ |
|
not modelled |
5.5 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative nad(p)h nitroreductase ydfn;
PDBTitle: crystal structure of putative nitroreductase ydfn (2632848) from2 bacillus subtilis at 1.65 a resolution
|
|
|
| 80 | c4ifdI_ |
|
not modelled |
5.5 |
56 |
PDB header:hydrolase/rna
Chain: I: PDB Molecule:exosome complex component csl4;
PDBTitle: crystal structure of an 11-subunit eukaryotic exosome complex bound to2 rna
|
|
|
| 81 | c1zcjA_ |
|
not modelled |
5.2 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peroxisomal bifunctional enzyme;
PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase
|
|
|