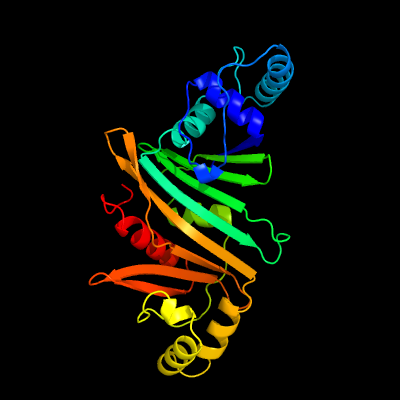
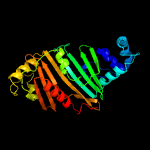
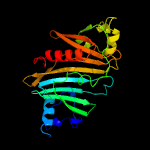
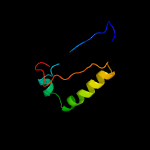
1 c5vbaA_
100.0
80
PDB header: chaperone, hydrolaseChain: A: PDB Molecule: lysozyme, esx-1 secretion-associated protein espg1 chimera;PDBTitle: structure of espg1 chaperone from the type vii (esx-1) secretion2 system determined with the assistance of n-terminal t4 lysozyme3 fusion
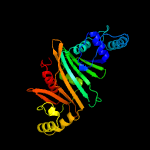
2 c4l4wB_
100.0
27
PDB header: protein transportChain: B: PDB Molecule: espg3;PDBTitle: structure of espg3 chaperone from the type vii (esx-3) secretion2 system
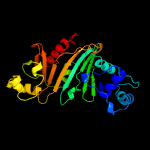
3 c4w4iA_
100.0
24
PDB header: protein transportChain: A: PDB Molecule: esx-3 secretion-associated protein espg3;PDBTitle: crystal structure of espg3 from the esx-3 type vii secretion system of2 m. tuberculosis
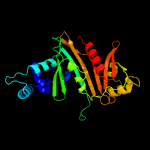
4 c4kxrC_
100.0
23
PDB header: protein transportChain: C: PDB Molecule: espg5;PDBTitle: structure of the mycobacterium tuberculosis type vii secretion system2 chaperone espg5 in complex with pe25-ppe41 dimer
5 c4rclB_
100.0
28
PDB header: chaperoneChain: B: PDB Molecule: espg3;PDBTitle: structure of espg3 chaperone from the type vii (esx-3) secretion2 system, space group p43212
6 c2l3aA_
19.6
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of homodimer protein sp_0782 (7-79) from2 streptococcus pneumoniae northeast structural genomics consortium3 target spr104 .
7 c5w4zA_
16.1
23
PDB header: flavoproteinChain: A: PDB Molecule: riboflavin lyase;PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
8 c3u5gK_
15.0
24
PDB header: ribosomeChain: K: PDB Molecule: 40s ribosomal protein s10-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome b
9 c2v4oB_
14.3
23
PDB header: hydrolaseChain: B: PDB Molecule: multifunctional protein sur e;PDBTitle: crystal structure of salmonella typhimurium sure at 2.752 angstrom resolution in monoclinic form
10 d1qnta1
13.3
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain
11 c3pm7A_
12.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ef_3132 protein from enterococcus faecalis at the2 resolution 2a, northeast structural genomics consortium target efr184
12 c4u5pA_
12.0
26
PDB header: isomeraseChain: A: PDB Molecule: rhcc;PDBTitle: crystal structure of native rhcc (yp_702633.1) from rhodococcus jostii2 rha1 at 1.78 angstrom
13 c2ltdA_
11.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ydbc;PDBTitle: solution nmr structure of apo ydbc from lactococcus lactis, northeast2 structural genomics consortium (nesg) target kr150
14 c2vxhF_
10.0
6
PDB header: oxidoreductaseChain: F: PDB Molecule: chlorite dismutase;PDBTitle: the crystal structure of chlorite dismutase: a detox enzyme2 producing molecular oxygen
15 d1g26a_
9.2
63
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Granulin repeatFamily: Granulin repeat
16 c3obhA_
8.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: x-ray crystal structure of protein sp_0782 (7-79) from streptococcus2 pneumoniae. northeast structural genomics consortium target spr104
17 c5craB_
8.7
43
PDB header: hydrolaseChain: B: PDB Molecule: sdea;PDBTitle: structure of the sdea dub domain
18 c3jcmK_
8.5
17
PDB header: transcriptionChain: K: PDB Molecule: u4/u6 small nuclear ribonucleoprotein prp3;PDBTitle: cryo-em structure of the spliceosomal u4/u6.u5 tri-snrnp
19 c2x5kO_
8.4
18
PDB header: oxidoreductaseChain: O: PDB Molecule: d-erythrose-4-phosphate dehydrogenase;PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
20 c4dibF_
8.3
18
PDB header: oxidoreductaseChain: F: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 bacillus anthracis str. sterne
21 c5jyfB_
not modelled
8.3
9
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structures of streptococcus agalactiae gbs gapdh in different2 enzymatic states
22 c5a13J_
not modelled
8.1
9
PDB header: oxidoreductaseChain: J: PDB Molecule: chlorite dismutase;PDBTitle: crystal structure of chlorite dismutase from2 magnetospirillum sp. in complex with thiocyanate
23 c2kimA_
not modelled
8.0
10
PDB header: transferaseChain: A: PDB Molecule: o6-methylguanine-dna methyltransferase;PDBTitle: 1.7-mm microcryoprobe solution nmr structure of an o6-methylguanine2 dna methyltransferase family protein from vibrio parahaemolyticus.3 northeast structural genomics consortium target vpr247.
24 c2i5pO_
not modelled
7.9
18
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
25 c4qx6A_
not modelled
7.8
9
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 streptococcus agalactiae nem316 at 2.46 angstrom resolution
26 c1obfO_
not modelled
7.6
18
PDB header: glycolytic pathwayChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.7 a3 resolution.
27 c5ur0B_
not modelled
7.5
18
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase2 from naegleria gruberi
28 c3hq4R_
not modelled
7.4
18
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
29 c1s7cA_
not modelled
7.4
18
PDB header: structural genomics, oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
30 c5ld5C_
not modelled
7.4
9
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of a bacterial dehydrogenase at 2.19 angstroms2 resolution
31 c2b4rQ_
not modelled
7.4
18
PDB header: oxidoreductaseChain: Q: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
32 c3c6vB_
not modelled
7.3
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable tautomerase/dehalogenase au4130;PDBTitle: crystal structure of au4130/apc7354, a probable enzyme from the2 thermophilic fungus aspergillus fumigatus
33 c3h9eO_
not modelled
7.3
18
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase, testis-specific;PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
34 c2ep7B_
not modelled
7.3
27
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
35 c3cieC_
not modelled
7.2
18
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
36 c1hdgO_
not modelled
7.2
27
PDB header: oxidoreductase (aldehy(d)-nad(a))Chain: O: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
37 c2d2iO_
not modelled
7.0
18
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
38 c1cerC_
not modelled
7.0
18
PDB header: oxidoreductase (aldehyde(d)-nad(a))Chain: C: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
39 c5j9gB_
not modelled
7.0
9
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-p dehydrogenase;PDBTitle: structure of lactobacillus acidophilus glyceraldehyde-3-phosphate2 dehydrogenase at 2.21 angstrom resolution
40 c3docD_
not modelled
6.9
18
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate dehydrogenase2 from brucella melitensis
41 c2pkrI_
not modelled
6.9
18
PDB header: oxidoreductaseChain: I: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase aor;PDBTitle: crystal structure of (a+cte)4 chimeric form of photosyntetic2 glyceraldehyde-3-phosphate dehydrogenase, complexed with nadp
42 d1ea0a3
not modelled
6.9
18
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases
43 c3b20R_
not modelled
6.9
18
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase (nadp+);PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase2 complexed with nadfrom synechococcus elongatus"
44 c1ihxD_
not modelled
6.8
18
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
45 c6ok4A_
not modelled
6.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase (gapdh)2 from chlamydia trachomatis with bound nad
46 c1rm4O_
not modelled
6.7
18
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
47 c2gfqC_
not modelled
6.7
24
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: upf0204 protein ph0006;PDBTitle: structure of protein of unknown function ph0006 from pyrococcus2 horikoshii
48 c1i32D_
not modelled
6.7
9
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
49 c3hjaB_
not modelled
6.7
18
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 borrelia burgdorferi
50 c4rdkB_
not modelled
6.3
21
PDB header: viral proteinChain: B: PDB Molecule: capsid;PDBTitle: crystal structure of norovirus boxer p domain in complex with lewis b2 tetrasaccharide
51 c3i4jC_
not modelled
6.3
16
PDB header: transferaseChain: C: PDB Molecule: aminotransferase, class iii;PDBTitle: crystal structure of aminotransferase, class iii from deinococcus2 radiodurans
52 c3sthA_
not modelled
6.3
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
53 d1l3ac_
not modelled
6.2
26
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: Plant transcriptional regulator PBF-2
54 d2glia4
not modelled
6.2
31
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
55 c4qyjD_
not modelled
6.0
15
PDB header: oxidoreductaseChain: D: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure of phenylacetaldehyde dehydrogenase from pseudomonas putida2 s12
56 c3gx4X_
not modelled
5.9
10
PDB header: dna binding protein/dnaChain: X: PDB Molecule: alkyltransferase-like protein 1;PDBTitle: crystal structure analysis of s. pombe atl in complex with dna
57 d1mgta1
not modelled
5.7
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain
58 c3h8dC_
not modelled
5.2
24
PDB header: motor protein/signaling proteinChain: C: PDB Molecule: myosin-vi;PDBTitle: crystal structure of myosin vi in complex with dab2 peptide
59 c4r8aF_
not modelled
5.2
21
PDB header: hydrolase/dnaChain: F: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of pafan1 - 5' flap dna complex