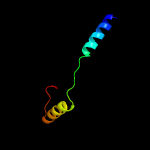
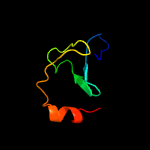
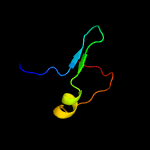
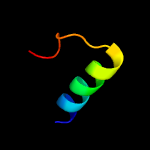
1 c4kk7A_
100.0
35
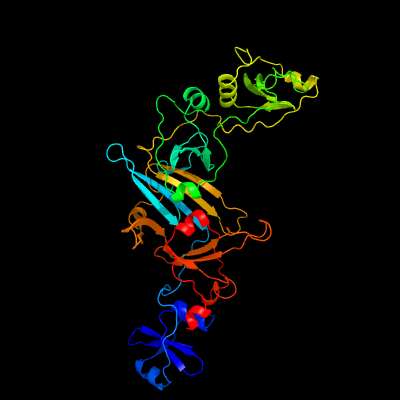
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion system protein eccb1;PDBTitle: structure of eccb1 from the type vii (esx-1) secretion system of2 mycobacterium tuberculosis.
2 c5cyuA_
100.0
35
PDB header: membrane proteinChain: A: PDB Molecule: conserved membrane protein;PDBTitle: structure of the soluble domain of eccb1 from the mycobacterium2 smegmatis esx-1 secretion system.
3 c6navI_
42.8
18
PDB header: structural proteinChain: I: PDB Molecule: m9ud72;PDBTitle: cryo-em reconstruction of sulfolobus islandicus lal14/1 pilus
4 d2fb5a1
36.8
19
Fold: YojJ-likeSuperfamily: YojJ-likeFamily: YojJ-like
5 c1k6nH_
30.3
22
PDB header: photosynthesisChain: H: PDB Molecule: photosynthetic reaction center h subunit;PDBTitle: e(l212)a,d(l213)a double mutant structure of photosynthetic reaction2 center from rhodobacter sphaeroides
6 d2i5nh1
30.3
20
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: Photosynthetic reaction centre, H-chain, cytoplasmic domain
7 c2i5nH_
23.6
19
PDB header: photosynthesisChain: H: PDB Molecule: reaction center protein h chain;PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
8 c2wmmA_
21.1
16
PDB header: cell cycleChain: A: PDB Molecule: chromosome partition protein mukb;PDBTitle: crystal structure of the hinge domain of mukb
9 d1rzhh1
18.5
22
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: Photosynthetic reaction centre, H-chain, cytoplasmic domain
10 c2ixsB_
17.4
11
PDB header: hydrolaseChain: B: PDB Molecule: sdai restriction endonuclease;PDBTitle: structure of sdai restriction endonuclease
11 c5xefA_
16.9
5
PDB header: chaperoneChain: A: PDB Molecule: flagellar protein flis;PDBTitle: crystal structure of flagellar chaperone from bacteria
12 d1vh6a_
14.3
19
Fold: Four-helical up-and-down bundleSuperfamily: Flagellar export chaperone FliSFamily: Flagellar export chaperone FliS
13 c1vh6A_
14.3
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flagellar protein flis;PDBTitle: crystal structure of a flagellar protein
14 c3k1iA_
13.3
10
PDB header: chaperoneChain: A: PDB Molecule: flagellar protein;PDBTitle: crystal strcture of flis-hp1076 complex in h. pylori
15 c5mrwL_
12.7
13
PDB header: hydrolaseChain: L: PDB Molecule: potassium-transporting atpase kdpf subunit;PDBTitle: structure of the kdpfabc complex
16 c6hrbD_
12.7
13
PDB header: membrane proteinChain: D: PDB Molecule: potassium-transporting atpase kdpf subunit;PDBTitle: cryo-em structure of the kdpfabc complex in an e2 inward-facing state2 (state 2)
17 c5mrwD_
12.7
13
PDB header: hydrolaseChain: D: PDB Molecule: potassium-transporting atpase kdpf subunit;PDBTitle: structure of the kdpfabc complex
18 c5mrwH_
12.7
13
PDB header: hydrolaseChain: H: PDB Molecule: potassium-transporting atpase kdpf subunit;PDBTitle: structure of the kdpfabc complex
19 c6hraD_
12.7
13
PDB header: membrane proteinChain: D: PDB Molecule: potassium-transporting atpase kdpf subunit;PDBTitle: cryo-em structure of the kdpfabc complex in an e1 outward-facing state2 (state 1)
20 d1y7ma1
12.1
15
Fold: L,D-transpeptidase catalytic domain-likeSuperfamily: L,D-transpeptidase catalytic domain-likeFamily: L,D-transpeptidase catalytic domain-like
21 c2ciuA_
not modelled
10.5
26
PDB header: protein transportChain: A: PDB Molecule: import inner membrane translocase subunit tim21PDBTitle: structure of the ims domain of the mitochondrial import2 protein tim21 from s. cerevisiae
22 d2f23a2
not modelled
10.0
45
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain
23 c2lf3A_
not modelled
9.2
56
PDB header: signaling proteinChain: A: PDB Molecule: effector protein hopab3;PDBTitle: solution nmr structure of hoppmal_281_385 from pseudomonas syringae2 pv. maculicola str. es4326, midwest center for structural genomics3 target apc40104.5 and northeast structural genomics consortium target4 pst2a
24 c3c1zA_
not modelled
9.1
13
PDB header: dna binding proteinChain: A: PDB Molecule: dna integrity scanning protein disa;PDBTitle: structure of the ligand-free form of a bacterial dna damage sensor2 protein
25 c3hshA_
not modelled
9.0
18
PDB header: protein bindingChain: A: PDB Molecule: collagen alpha-1(xviii) chain;PDBTitle: crystal structure of human collagen xviii trimerization domain2 (tetragonal crystal form)
26 c6cfwE_
not modelled
8.9
11
PDB header: membrane proteinChain: E: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
27 c6ch3B_
not modelled
8.1
23
PDB header: structural proteinChain: B: PDB Molecule: flagellar secretion chaperone flis,flagellin;PDBTitle: crystal structure of the cytoplasmic domain of flha and flis-flic2 complex
28 c6gyyB_
not modelled
7.6
21
PDB header: transferaseChain: B: PDB Molecule: diadenylate cyclase;PDBTitle: crystal structure of daca from staphylococcus aureus, n166c/t172c2 double mutant
29 c3n3fB_
not modelled
7.6
21
PDB header: protein bindingChain: B: PDB Molecule: collagen alpha-1(xv) chain;PDBTitle: crystal structure of the human collagen xv trimerization domain: a2 potent trimerizing unit common to multiplexin collagens
30 c2l02B_
not modelled
7.6
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein bt2368 from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr375
31 d1vqou1
not modelled
7.3
18
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Ribosomal protein L24e
32 c3ccjU_
not modelled
7.3
18
PDB header: ribosomeChain: U: PDB Molecule: 50s ribosomal protein l24e;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation c2534u
33 c3bmbB_
not modelled
7.1
29
PDB header: rna binding proteinChain: B: PDB Molecule: regulator of nucleoside diphosphate kinase;PDBTitle: crystal structure of a new rna polymerase interacting2 protein
34 c6f0kA_
not modelled
7.0
18
PDB header: membrane proteinChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: alternative complex iii
35 d1ixsa_
not modelled
6.6
30
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain
36 c2p4vA_
not modelled
6.0
27
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
37 d2zjru1
not modelled
5.9
20
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L28
38 d2b3ga1
not modelled
5.9
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB
39 c2kncA_
not modelled
5.9
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
40 c2a7oA_
not modelled
5.8
15
PDB header: transcriptionChain: A: PDB Molecule: huntingtin interacting protein b;PDBTitle: solution structure of the hset2/hypb sri domain
41 c2k9yA_
not modelled
5.7
3
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
42 c2k9yB_
not modelled
5.7
3
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
43 c4jonA_
not modelled
5.5
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: centrosomal protein of 170 kda;PDBTitle: crystal structure of a centrosomal protein 170kda, transcript variant2 beta (cep170) from homo sapiens at 2.15 a resolution (psi community3 target, sundstrom)
44 c5dn6P_
not modelled
5.5
18
PDB header: hydrolaseChain: P: PDB Molecule: atp synthase f0 subcomplex c subunit;PDBTitle: atp synthase from paracoccus denitrificans
45 d2etna2
not modelled
5.3
42
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain
46 c2mhgA_
not modelled
5.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of protein np_254181.1 from pseudomonas aeruginosa pa01
47 c2o35A_
not modelled
5.1
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf1244;PDBTitle: protein of unknown function (duf1244) from sinorhizobium meliloti
48 d2o35a1
not modelled
5.1
26
Fold: SMc04008-likeSuperfamily: SMc04008-likeFamily: SMc04008-like
49 c2etnA_
not modelled
5.1
42
PDB header: transcriptionChain: A: PDB Molecule: anti-cleavage anti-grea transcription factorPDBTitle: crystal structure of thermus aquaticus gfh1
50 c2zkru_
not modelled
5.0
35
PDB header: ribosomal protein/rnaChain: U: PDB Molecule: rna expansion segment es41;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map