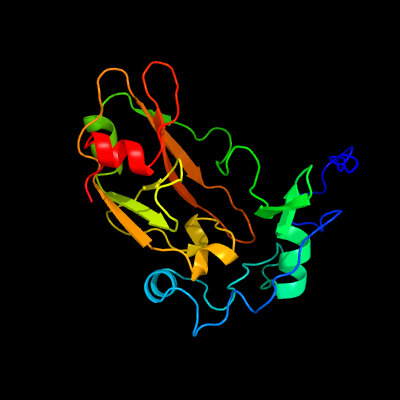
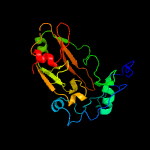
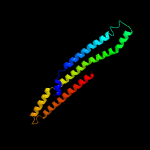
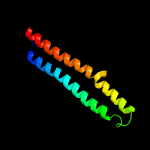
1 c4qlpB_
100.0
98
PDB header: hydrolase/protein bindingChain: B: PDB Molecule: alanine and proline rich protein, tuberculosis necrotizingPDBTitle: atomic structure of tuberculosis necrotizing toxin (tnt) complexed2 with its immunity factor ift
2 c2g38B_
97.0
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
3 d2g38b1
97.0
13
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
4 c4iogD_
95.6
20
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
5 c5xfsB_
94.0
14
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
6 c3zbhC_
93.6
8
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
7 c2vs0B_
93.4
12
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
8 c3gvmA_
93.2
15
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 d1wa8a1
93.0
18
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
10 c4lwsA_
91.1
13
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
11 d1eq1a_
82.5
13
Fold: Apolipophorin-IIISuperfamily: Apolipophorin-IIIFamily: Apolipophorin-III
12 c4b1vN_
69.6
73
PDB header: structural proteinChain: N: PDB Molecule: phosphatase and actin regulator 1;PDBTitle: structure of the phactr1 rpel-n domain bound to g-actin
13 c4b1vM_
69.6
73
PDB header: structural proteinChain: M: PDB Molecule: phosphatase and actin regulator 1;PDBTitle: structure of the phactr1 rpel-n domain bound to g-actin
14 c4b1uM_
67.7
73
PDB header: structural proteinChain: M: PDB Molecule: phosphatase and actin regulator 1;PDBTitle: structure of the phactr1 rpel domain and rpel motif directed2 assemblies with g-actin reveal the molecular basis for actin binding3 cooperativity.
15 c4lwsB_
67.2
14
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
16 c2kg7B_
64.7
10
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
17 d1g4us1
54.0
11
Fold: Four-helical up-and-down bundleSuperfamily: Bacterial GAP domainFamily: Bacterial GAP domain
18 d1wa8b1
49.5
5
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
19 c4wj2A_
46.7
11
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
20 c6b7nC_
45.0
10
PDB header: viral proteinChain: C: PDB Molecule: spike protein;PDBTitle: cryo-electron microscopy structure of porcine delta coronavirus spike2 protein in the pre-fusion state
21 c4xy3A_
not modelled
40.1
13
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
22 c3mhsE_
not modelled
20.7
15
PDB header: hydrolase/transcription regulator/proteiChain: E: PDB Molecule: saga-associated factor 73;PDBTitle: structure of the saga ubp8/sgf11/sus1/sgf73 dub module bound to2 ubiquitin aldehyde
23 c2mn4A_
not modelled
20.4
70
PDB header: de novo proteinChain: A: PDB Molecule: computational designed protein based on structure templatePDBTitle: nmr solution structure of a computational designed protein based on2 structure template 1cy5
24 c1rh1A_
not modelled
19.6
12
PDB header: antibioticChain: A: PDB Molecule: colicin b;PDBTitle: crystal structure of the cytotoxic bacterial protein2 colicin b at 2.5 a resolution
25 c6cv0C_
not modelled
19.2
14
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: cryo-electron microscopy structure of infectious bronchitis2 coronavirus spike protein
26 c6nzkB_
not modelled
19.2
15
PDB header: viral proteinChain: B: PDB Molecule: spike surface glycoprotein;PDBTitle: structural basis for human coronavirus attachment to sialic acid2 receptors
27 c4i0xA_
not modelled
16.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
28 c2vxzA_
not modelled
16.5
7
PDB header: viral proteinChain: A: PDB Molecule: pyrsv_gp04;PDBTitle: crystal structure of hypothetical protein pyrsv_gp04 from pyrobaculum2 spherical virus
29 c2lo3A_
not modelled
14.4
15
PDB header: transcriptionChain: A: PDB Molecule: saga-associated factor 73;PDBTitle: solution structure of sgf73(59-102) zinc finger domain
30 c1qu7A_
not modelled
12.9
6
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: four helical-bundle structure of the cytoplasmic domain of a serine2 chemotaxis receptor
31 c2zp2B_
not modelled
12.6
13
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
32 c3jclC_
not modelled
12.5
13
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: cryo-electron microscopy structure of a coronavirus spike glycoprotein2 trimer
33 c5x5bB_
not modelled
12.2
15
PDB header: viral proteinChain: B: PDB Molecule: spike glycoprotein;PDBTitle: prefusion structure of sars-cov spike glycoprotein, conformation 2
34 c3g67A_
not modelled
11.9
5
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
35 c5x5fC_
not modelled
11.6
10
PDB header: viral proteinChain: C: PDB Molecule: s protein;PDBTitle: prefusion structure of mers-cov spike glycoprotein, conformation 2
36 c5i08A_
not modelled
10.9
15
PDB header: viral proteinChain: A: PDB Molecule: spike glycoprotein, envelope glycoprotein chimera;PDBTitle: prefusion structure of a human coronavirus spike protein
37 c5xlrC_
not modelled
10.7
15
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: structure of sars-cov spike glycoprotein
38 c1ei3E_
not modelled
10.3
5
PDB header: blood clottingChain: E: PDB Molecule: fibrinogen;PDBTitle: crystal structure of native chicken fibrinogen
39 c5o9eB_
not modelled
10.3
41
PDB header: ribosomeChain: B: PDB Molecule: putative u3 small nucleolar ribonucleoprotein protein;PDBTitle: crystal structure of the imp4-mpp10 complex from chaetomium2 thermophilum
40 c6nb3B_
not modelled
9.6
10
PDB header: virusChain: B: PDB Molecule: spike glycoprotein;PDBTitle: mers-cov complex with human neutralizing lca60 antibody fab fragment2 (state 1)
41 c3ghgK_
not modelled
9.5
8
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
42 c5szsC_
not modelled
9.1
14
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: glycan shield and epitope masking of a coronavirus spike protein2 observed by cryo-electron microscopy
43 c4i0xJ_
not modelled
9.1
9
PDB header: structural genomics, unknown functionChain: J: PDB Molecule: esat-6-like protein mab_3113;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
44 d2gqba1
not modelled
9.1
17
Fold: RPA2825-likeSuperfamily: RPA2825-likeFamily: RPA2825-like
45 c5wrgB_
not modelled
8.5
15
PDB header: virus like particleChain: B: PDB Molecule: spike glycoprotein;PDBTitle: sars-cov spike glycoprotein
46 d1he1a_
not modelled
8.5
9
Fold: Four-helical up-and-down bundleSuperfamily: Bacterial GAP domainFamily: Bacterial GAP domain
47 c6n1fD_
not modelled
8.2
22
PDB header: oxidoreductaseChain: D: PDB Molecule: oxidoreductase, 2og-fe(ii) oxygenase family;PDBTitle: crystal structure of oxidoreductase, 2og-fe(ii) oxygenase family, from2 burkholderia pseudomallei
48 c2ieqC_
not modelled
8.1
14
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: core structure of s2 from the human coronavirus nl63 spike2 glycoprotein
49 c3qflA_
not modelled
8.0
13
PDB header: protein bindingChain: A: PDB Molecule: mla10;PDBTitle: coiled-coil domain-dependent homodimerization of intracellular mla2 immune receptors defines a minimal functional module for triggering3 cell death
50 c6akmA_
not modelled
8.0
13
PDB header: protein bindingChain: A: PDB Molecule: suppressor of ikbke 1;PDBTitle: crystal structure of slmap-sike1 complex
51 c2jv4A_
not modelled
7.9
45
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis/trans isomerase;PDBTitle: structure characterisation of pina ww domain and comparison2 with other group iv ww domains, pin1 and ess1
52 c6cs2A_
not modelled
7.4
15
PDB header: viral protein/hydrolaseChain: A: PDB Molecule: spike glycoprotein,fibritin;PDBTitle: sars spike glycoprotein - human ace2 complex, stabilized variant, all2 ace2-bound particles
53 c6dmaD_
not modelled
6.8
19
PDB header: de novo proteinChain: D: PDB Molecule: dhd15_closed_b;PDBTitle: dhd15_closed
54 c3lnrA_
not modelled
6.8
16
PDB header: signaling proteinChain: A: PDB Molecule: aerotaxis transducer aer2;PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
55 c4e40A_
not modelled
6.7
12
PDB header: transport proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the haptoglobin-hemoglobin receptor of trypanosoma congolense
56 d1khia2
not modelled
6.6
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
57 c4ut1A_
not modelled
6.4
17
PDB header: motor proteinChain: A: PDB Molecule: flagellar hook-associated protein;PDBTitle: the structure of the flagellar hook junction protein flgk2 from burkholderia pseudomallei
58 c1sfcD_
not modelled
6.3
9
PDB header: transport proteinChain: D: PDB Molecule: protein (snap-25b);PDBTitle: neuronal synaptic fusion complex
59 c1deqO_
not modelled
6.3
6
PDB header: blood clottingChain: O: PDB Molecule: fibrinogen (beta chain);PDBTitle: the crystal structure of modified bovine fibrinogen (at ~42 angstrom resolution)
60 c2d4yA_
not modelled
6.1
12
PDB header: structural proteinChain: A: PDB Molecule: flagellar hook-associated protein 1;PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
61 c3ivrA_
not modelled
6.1
21
PDB header: ligaseChain: A: PDB Molecule: putative long-chain-fatty-acid coa ligase;PDBTitle: crystal structure of putative long-chain-fatty-acid coa ligase from2 rhodopseudomonas palustris cga009
62 d1owaa_
not modelled
6.1
7
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat
63 c3gw6F_
not modelled
6.0
14
PDB header: chaperoneChain: F: PDB Molecule: endo-n-acetylneuraminidase;PDBTitle: intramolecular chaperone
64 d1fx7a3
not modelled
6.0
35
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like
65 c1ei3C_
not modelled
6.0
9
PDB header: blood clottingChain: C: PDB Molecule: fibrinogen;PDBTitle: crystal structure of native chicken fibrinogen
66 c6ozvA_
not modelled
5.9
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: txo1;PDBTitle: the structure of condensation and adenylation domains of teixobactin-2 producing nonribosomal peptide synthetase txo1 serine module in3 complex with amp
67 c2i56A_
not modelled
5.8
15
PDB header: isomerase, metal-binding proteinChain: A: PDB Molecule: l-rhamnose isomerase;PDBTitle: crystal structure of l-rhamnose isomerase from pseudomonas2 stutzeri with l-rhamnose
68 c2choA_
not modelled
5.7
13
PDB header: hydrolaseChain: A: PDB Molecule: glucosaminidase;PDBTitle: bacteroides thetaiotaomicron hexosaminidase with o-2 glcnacase activity
69 c4nl6C_
not modelled
5.7
15
PDB header: splicingChain: C: PDB Molecule: survival motor neuron protein;PDBTitle: structure of the full-length form of the protein smn found in healthy2 patients
70 d1z6ra1
not modelled
5.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ROK associated domain
71 d2p0va1
not modelled
5.5
20
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: CPF0428-like
72 c2p0vA_
not modelled
5.5
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein bt3781;PDBTitle: crystal structure of bt3781 protein from bacteroides thetaiotaomicron,2 northeast structural genomics target btr58
73 c2l0gA_
not modelled
5.4
33
PDB header: protein bindingChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution nmr structure of ubiquitin-binding motif (ubm2) of human2 polymerase iota
74 d1lm5a_
not modelled
5.3
9
Fold: beta-hairpin-alpha-hairpin repeatSuperfamily: Plakin repeatFamily: Plakin repeat
75 d1szia_
not modelled
5.3
13
Fold: Four-helical up-and-down bundleSuperfamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domainFamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domain
76 c2k4vA_
not modelled
5.1
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pa1076;PDBTitle: solution structure of uncharacterized protein pa1076 from pseudomonas2 aeruginosa. northeast structural genomics consortium (nesg) target3 pat3, ontario center for structural proteomics target pa1076 .
77 c4qmdB_
not modelled
5.1
9
PDB header: cell adhesionChain: B: PDB Molecule: envoplakin;PDBTitle: crystal structure of human envoplakin plakin repeat domain
78 c2n03A_
not modelled
5.0
15
PDB header: structural proteinChain: A: PDB Molecule: plectin;PDBTitle: solution nmr structure plectin repeat domain 6 (4403-4606) of plectin2 from homo sapiens, northeast structural genomics consortium (nesg)3 target hr6354e