| 1 |
|
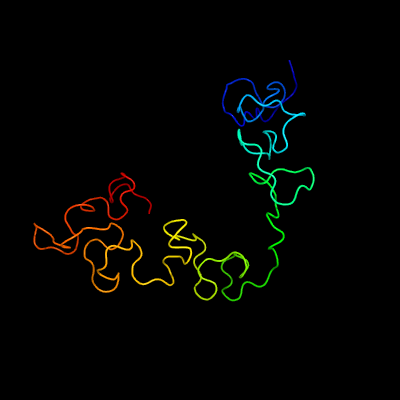
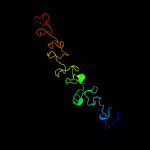
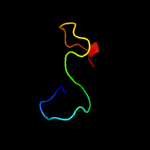
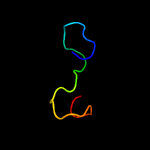
PDB 3fby chain C
Region: 14 - 169
Aligned: 152
Modelled: 156
Confidence: 99.9%
Identity: 26%
PDB header:cell adhesion
Chain: C: PDB Molecule:cartilage oligomeric matrix protein;
PDBTitle: the crystal structure of the signature domain of cartilage oligomeric2 matrix protein.
Phyre2
| 2 |
|
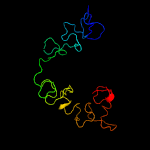
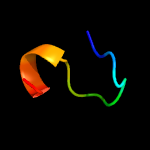
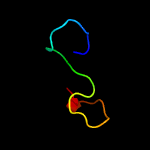
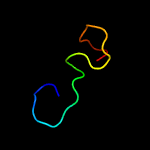
PDB 1yo8 chain A
Region: 16 - 169
Aligned: 150
Modelled: 154
Confidence: 99.9%
Identity: 25%
PDB header:cell adhesion
Chain: A: PDB Molecule:thrombospondin-2;
PDBTitle: structure of the c-terminal domain of human thrombospondin-2
Phyre2
| 3 |
|
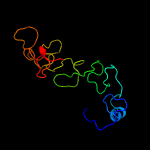
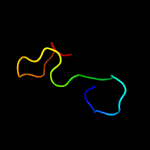
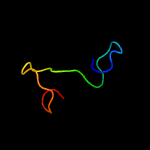
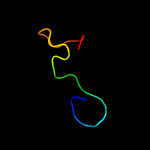
PDB 1ux6 chain A
Region: 54 - 169
Aligned: 108
Modelled: 116
Confidence: 99.8%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:thrombospondin-1;
PDBTitle: structure of a thrombospondin c-terminal fragment reveals a novel2 calcium core in the type 3 repeats
Phyre2
| 4 |
|
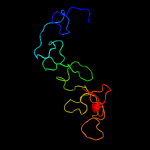
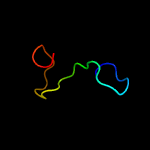
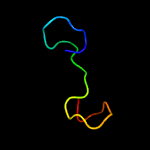
PDB 1ux6 chain A domain 2
Region: 17 - 169
Aligned: 106
Modelled: 107
Confidence: 99.7%
Identity: 25%
Fold: TSP type-3 repeat
Superfamily: TSP type-3 repeat
Family: TSP type-3 repeat
Phyre2
| 5 |
|
PDB 5wtl chain B
Region: 22 - 168
Aligned: 122
Modelled: 137
Confidence: 99.3%
Identity: 25%
PDB header:membrane protein
Chain: B: PDB Molecule:ompa family protein;
PDBTitle: crystal structure of the periplasmic portion of outer membrane protein2 a (ompa) from capnocytophaga gingivalis
Phyre2
| 6 |
|
PDB 1tzo chain A
Region: 21 - 34
Aligned: 14
Modelled: 14
Confidence: 63.8%
Identity: 36%
Fold: Anthrax protective antigen
Superfamily: Anthrax protective antigen
Family: Anthrax protective antigen
Phyre2
| 7 |
|
PDB 1acc chain A
Region: 18 - 34
Aligned: 17
Modelled: 17
Confidence: 38.7%
Identity: 29%
Fold: Anthrax protective antigen
Superfamily: Anthrax protective antigen
Family: Anthrax protective antigen
Phyre2
| 8 |
|
PDB 1ohz chain B
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 26.8%
Identity: 42%
Fold: Type I dockerin domain
Superfamily: Type I dockerin domain
Family: Type I dockerin domain
Phyre2
| 9 |
|
PDB 2j42 chain A
Region: 16 - 34
Aligned: 19
Modelled: 19
Confidence: 26.5%
Identity: 26%
PDB header:toxin
Chain: A: PDB Molecule:c2 toxin component-ii;
PDBTitle: low quality crystal structure of the transport component c2-2 ii of the c2-toxin from clostridium botulinum
Phyre2
| 10 |
|
PDB 4dh2 chain B
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 23.8%
Identity: 31%
PDB header:cell adhesion/protein binding
Chain: B: PDB Molecule:dockerin type 1;
PDBTitle: crystal structure of coh-olpc(cthe_0452)-doc435(cthe_0435) complex: a2 novel type i cohesin-dockerin complex from clostridium thermocellum3 attc 27405
Phyre2
| 11 |
|
PDB 3p0d chain D
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 21.7%
Identity: 42%
PDB header:hydrolase
Chain: D: PDB Molecule:glycoside hydrolase family 9;
PDBTitle: crystal structure of a multimodular ternary protein complex from2 clostridium thermocellum
Phyre2
| 12 |
|
PDB 4uyq chain B
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 21.2%
Identity: 42%
PDB header:cell adhesion/protein binding
Chain: B: PDB Molecule:cellulosomal scaffoldin adaptor protein b;
PDBTitle: high resolution structure of the third cohesin scac in complex with2 the scab dockerin with a mutation in the c-terminal helix (in to si)3 from acetivibrio cellulolyticus displaying a type i interaction.
Phyre2
| 13 |
|
PDB 2y3n chain B
Region: 139 - 167
Aligned: 29
Modelled: 29
Confidence: 20.1%
Identity: 28%
PDB header:structrual protein/hydrolase
Chain: B: PDB Molecule:cellulosomal family-48 processive glycoside hydrolase;
PDBTitle: type ii cohesin-dockerin domain from bacteroides cellolosolvens
Phyre2
| 14 |
|
PDB 3ul4 chain B
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 16.2%
Identity: 42%
PDB header:cell adhesion/protein binding
Chain: B: PDB Molecule:cellulosome enzyme, dockerin type i;
PDBTitle: crystal structure of coh-olpa(cthe_3080)-doc918(cthe_0918) complex: a2 novel type i cohesin-dockerin complex from clostridium thermocellum3 attc 27405
Phyre2
| 15 |
|
PDB 1dav chain A
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 15.9%
Identity: 31%
Fold: Type I dockerin domain
Superfamily: Type I dockerin domain
Family: Type I dockerin domain
Phyre2
| 16 |
|
PDB 5nrm chain B
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 15.6%
Identity: 42%
PDB header:cell adhesion
Chain: B: PDB Molecule:doccel5: type i dockerin repeat domain from a.
PDBTitle: crystal structure of the sixth cohesin from acetivibrio2 cellulolyticus' scaffoldin b in complex with cel5 dockerin s51i, l52n3 mutant
Phyre2
| 17 |
|
PDB 5lxv chain B
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 15.2%
Identity: 50%
PDB header:protein binding
Chain: B: PDB Molecule:carbohydrate-binding protein wp_009985128;
PDBTitle: crystal structure of ruminococcus flavefaciens scaffoldin c cohesin in2 complex with a dockerin from an uncharacterized cbm-containing3 protein
Phyre2
| 18 |
|
PDB 2ccl chain B domain 1
Region: 143 - 168
Aligned: 26
Modelled: 26
Confidence: 15.1%
Identity: 46%
Fold: Type I dockerin domain
Superfamily: Type I dockerin domain
Family: Type I dockerin domain
Phyre2
| 19 |
|
PDB 2vn5 chain B
Region: 143 - 167
Aligned: 25
Modelled: 25
Confidence: 11.8%
Identity: 28%
PDB header:cell adhesion
Chain: B: PDB Molecule:endoglucanase a;
PDBTitle: the clostridium cellulolyticum dockerin displays a dual2 binding mode for its cohesin partner
Phyre2
| 20 |
|
PDB 2b59 chain B
Region: 143 - 167
Aligned: 25
Modelled: 25
Confidence: 10.2%
Identity: 16%
PDB header:hydrolase/structural protein
Chain: B: PDB Molecule:cellulosomal scaffolding protein a;
PDBTitle: the type ii cohesin dockerin complex
Phyre2