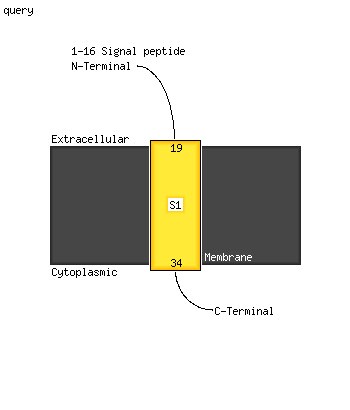
1 c5lc5q_
35.0
25
PDB header: oxidoreductaseChain: Q: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 4,PDBTitle: structure of mammalian respiratory complex i, class2
2 c4nqkD_
30.3
12
PDB header: hydrolase/apoptosisChain: D: PDB Molecule: probable atp-dependent rna helicase ddx58;PDBTitle: structure of an ubiquitin complex
3 c3zj1A_
20.4
44
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein nab2;PDBTitle: structure of nab2p tandem zinc finger 12
4 c3oruA_
18.6
32
PDB header: metal binding proteinChain: A: PDB Molecule: duf1989 family protein;PDBTitle: crystal structure of a duf1989 family protein (tm1040_0329) from2 silicibacter sp. tm1040 at 1.11 a resolution
5 c6e8dA_
17.8
36
PDB header: dna binding proteinChain: A: PDB Molecule: beta sliding clamp,dna mismatch repair protein mutl;PDBTitle: crystal structure of the bacillus subtilis sliding clamp-mutl complex.
6 c5i0cA_
17.6
21
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein yjdj;PDBTitle: crystal structure of predicted acyltransferase yjdj with acyl-coa n-2 acyltransferase domain from escherichia coli str. k-12
7 c6ekrA_
17.4
38
PDB header: hydrolaseChain: A: PDB Molecule: type ii site-specific deoxyribonuclease;PDBTitle: crystal structure of type iip restriction endonuclease kpn2i
8 c5b37A_
15.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: tryptophan dehydrogenase;PDBTitle: crystal structure of l-tryptophan dehydrogenase from nostoc2 punctiforme
9 c1cdlH_
14.9
50
PDB header: calcium-binding proteinChain: H: PDB Molecule: calcium/calmodulin-dependent protein kinase typePDBTitle: target enzyme recognition by calmodulin: 2.4 angstroms2 structure of a calmodulin-peptide complex
10 c5jthB_
14.8
50
PDB header: transferaseChain: B: PDB Molecule: myosin light chain kinase, smooth muscle;PDBTitle: crystal structure of e67a calmodulin - cam:rm20 complex
11 c5jqaB_
14.8
50
PDB header: calcium binding protein/protein bindingChain: B: PDB Molecule: myosin light chain kinase, smooth muscle;PDBTitle: cam:rm20 complex
12 c1cdlE_
14.8
50
PDB header: calcium-binding proteinChain: E: PDB Molecule: calcium/calmodulin-dependent protein kinase typePDBTitle: target enzyme recognition by calmodulin: 2.4 angstroms2 structure of a calmodulin-peptide complex
13 c1cdlF_
14.8
50
PDB header: calcium-binding proteinChain: F: PDB Molecule: calcium/calmodulin-dependent protein kinase typePDBTitle: target enzyme recognition by calmodulin: 2.4 angstroms2 structure of a calmodulin-peptide complex
14 c1cdlG_
14.7
50
PDB header: calcium-binding proteinChain: G: PDB Molecule: calcium/calmodulin-dependent protein kinase typePDBTitle: target enzyme recognition by calmodulin: 2.4 angstroms2 structure of a calmodulin-peptide complex
15 d3blha1
14.7
20
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Protein kinases, catalytic subunit
16 c2cseB_
14.5
20
PDB header: virusChain: B: PDB Molecule: major outer-capsid protein mu1;PDBTitle: features of reovirus outer-capsid protein mu1 revealed by2 electron and image reconstruction of the virion at 7.0-a3 resolution
17 c1qtxB_
14.5
50
PDB header: signaling proteinChain: B: PDB Molecule: protein (rs20);PDBTitle: the 1.65 angstrom structure of calmodulin rs20 peptide2 complex
18 c2o5gB_
14.3
50
PDB header: metal binding proteinChain: B: PDB Molecule: smooth muscle myosin light chain kinase peptide;PDBTitle: calmodulin-smooth muscle light chain kinase peptide complex
19 c1qs7B_
13.8
50
PDB header: metal binding protein/peptideChain: B: PDB Molecule: rs20;PDBTitle: the 1.8 angstrom structure of calmodulin rs20 peptide2 complex
20 c1vrkB_
13.7
50
PDB header: complex(calcium-binding protein/peptide)Chain: B: PDB Molecule: rs20;PDBTitle: the 1.9 angstrom structure of e84k-calmodulin rs20 peptide2 complex
21 c1qs7D_
not modelled
12.6
50
PDB header: metal binding protein/peptideChain: D: PDB Molecule: rs20;PDBTitle: the 1.8 angstrom structure of calmodulin rs20 peptide2 complex
22 c1jmuD_
not modelled
12.4
23
PDB header: viral proteinChain: D: PDB Molecule: protein mu-1;PDBTitle: crystal structure of the reovirus mu1/sigma3 complex
23 c2lweA_
not modelled
10.8
11
PDB header: signaling proteinChain: A: PDB Molecule: probable atp-dependent rna helicase ddx58;PDBTitle: solution structure of mutant (t170e) second card of human rig-i
24 d2j0141
not modelled
10.5
43
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p
25 c2j034_
not modelled
10.5
43
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
26 c2n5xA_
not modelled
10.0
44
PDB header: chaperoneChain: A: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: c-terminal domain of cdc37 cochaperone
27 c3bbo1_
not modelled
9.1
29
PDB header: ribosomeChain: 1: PDB Molecule: ribosomal protein l31;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
28 c3k8zD_
not modelled
8.2
30
PDB header: oxidoreductaseChain: D: PDB Molecule: nad-specific glutamate dehydrogenase;PDBTitle: crystal structure of gudb1 a decryptified secondary glutamate2 dehydrogenase from b. subtilis
29 c5c6gA_
not modelled
7.9
12
PDB header: cell cycleChain: A: PDB Molecule: agr133cp;PDBTitle: structural insights into the scc2-scc4 cohesin loader
30 c3k1qP_
not modelled
7.8
21
31 c6dzpg_
not modelled
7.6
29
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l6;PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 50s ribosomal2 subunit
32 c2nx6A_
not modelled
7.5
50
PDB header: structural proteinChain: A: PDB Molecule: nematocyst outer wall antigen;PDBTitle: structure of nowa cysteine rich domain 6
33 c2b664_
not modelled
7.5
29
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf1,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400
34 d1vs6z1
not modelled
6.8
43
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p
35 c3zj2A_
not modelled
6.8
50
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein nab2;PDBTitle: structure of nab2p tandem zinc finger 34
36 c1q68A_
not modelled
6.8
42
PDB header: membrane protein/transferaseChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: solution structure of t-cell surface glycoprotein cd4 and2 proto-oncogene tyrosine-protein kinase lck fragments
37 d1bgva2
not modelled
6.8
27
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases
38 c6nuwD_
not modelled
6.7
10
PDB header: cell cycleChain: D: PDB Molecule: inner kinetochore subunit ctf19;PDBTitle: yeast ctf19 complex
39 d1shyb2
not modelled
6.6
42
Fold: Trefoil/Plexin domain-likeSuperfamily: Plexin repeatFamily: Plexin repeat
40 c2hgj3_
not modelled
6.6
43
PDB header: ribosomeChain: 3: PDB Molecule: 50s ribosomal protein l31;PDBTitle: crystal structure of the 70s thermus thermophilus ribosome showing how2 the 16s 3'-end mimicks mrna e and p codons. this entry 2hgj contains3 50s ribosomal subunit. the 30s ribosomal subunit can be found in pdb4 entry 2hgi.
41 c5o60g_
not modelled
5.9
43
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l6;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
42 c1bvuF_
not modelled
5.8
27
PDB header: oxidoreductaseChain: F: PDB Molecule: protein (glutamate dehydrogenase);PDBTitle: glutamate dehydrogenase from thermococcus litoralis
43 c2mj2A_
not modelled
5.5
31
PDB header: viral proteinChain: A: PDB Molecule: agnoprotein;PDBTitle: structure of the dimerization domain of the human polyoma, jc virus2 agnoprotein is an amphipathic alpha-helix.
44 d1ut7a_
not modelled
5.3
50
Fold: NAC domainSuperfamily: NAC domainFamily: NAC domain
45 c2tmgD_
not modelled
5.1
33
PDB header: oxidoreductaseChain: D: PDB Molecule: protein (glutamate dehydrogenase);PDBTitle: thermotoga maritima glutamate dehydrogenase mutant s128r,2 t158e, n117r, s160e
46 c5n4cG_
not modelled
5.0
40
PDB header: hydrolaseChain: G: PDB Molecule: alpha-amanitin proprotein;PDBTitle: prolyl oligopeptidase b from galerina marginata bound to 35mer2 hydrolysis and macrocyclization substrate - s577a mutant
47 c3aoeC_
not modelled
5.0
30
PDB header: oxidoreductaseChain: C: PDB Molecule: glutamate dehydrogenase;PDBTitle: crystal structure of hetero-hexameric glutamate dehydrogenase from2 thermus thermophilus (leu bound form)