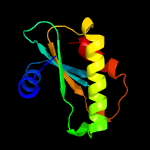
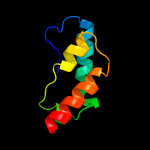
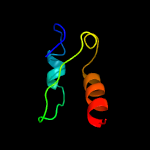
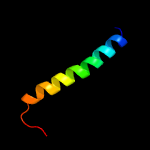
1 d1d6ta_
100.0
20
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: RNase P protein
2 d1a6fa_
100.0
26
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: RNase P protein
3 c2ljpA_
100.0
26
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease p protein component;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for e.coli2 ribonuclease p protein
4 d1nz0a_
100.0
19
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: RNase P protein
5 c4oxpA_
95.0
12
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease e;PDBTitle: x-ray crystal structure of the s1 and 5'-sensor domains of rnase e2 from caulobacter crescentus
6 c2c4rL_
52.4
13
PDB header: hydrolaseChain: L: PDB Molecule: ribonuclease e;PDBTitle: catalytic domain of e. coli rnase e
7 c3b0zA_
16.7
19
PDB header: protein transportChain: A: PDB Molecule: flagellar biosynthetic protein flhb;PDBTitle: crystal structure of cytoplasmic domain of flhb from salmonella2 typhimurium
8 c5aycA_
16.2
9
PDB header: transferaseChain: A: PDB Molecule: 4-o-beta-d-mannosyl-d-glucose phosphorylase;PDBTitle: crystal structure of ruminococcus albus 4-o-beta-d-mannosyl-d-glucose2 phosphorylase (ramp1) in complexes with sulfate and 4-o-beta-d-3 mannosyl-d-glucose
9 c2ml9A_
10.7
16
PDB header: membrane proteinChain: A: PDB Molecule: yop proteins translocation protein u;PDBTitle: solution structure of yscucn in a micellar complex with sds
10 c3b1sC_
9.9
32
PDB header: protein transportChain: C: PDB Molecule: flagellar biosynthetic protein flhb;PDBTitle: crystal structure of the cytoplasmic domain of flhb from aquifex2 aeolicus
11 c3b1sA_
9.9
32
PDB header: protein transportChain: A: PDB Molecule: flagellar biosynthetic protein flhb;PDBTitle: crystal structure of the cytoplasmic domain of flhb from aquifex2 aeolicus
12 c3qc3B_
9.4
12
PDB header: isomeraseChain: B: PDB Molecule: d-ribulose-5-phosphate-3-epimerase;PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
13 c3b1sE_
9.1
32
PDB header: protein transportChain: E: PDB Molecule: flagellar biosynthetic protein flhb;PDBTitle: crystal structure of the cytoplasmic domain of flhb from aquifex2 aeolicus
14 d1vpza_
9.0
27
Fold: CsrA-likeSuperfamily: CsrA-likeFamily: CsrA-like
15 c2jppB_
8.6
27
PDB header: translation/rnaChain: B: PDB Molecule: translational repressor;PDBTitle: structural basis of rsma/csra rna recognition: structure of2 rsme bound to the shine-dalgarno sequence of hcna mrna
16 c1vpzB_
8.5
27
PDB header: rna binding proteinChain: B: PDB Molecule: carbon storage regulator homolog;PDBTitle: crystal structure of a putative carbon storage regulator protein2 (csra, pa0905) from pseudomonas aeruginosa at 2.05 a resolution
17 c5dmbD_
8.3
19
PDB header: translationChain: D: PDB Molecule: carbon storage regulator homolog;PDBTitle: crystal structure of a translational regulator bound to a flagellar2 assembly factor
18 c5usrB_
8.1
19
PDB header: transferaseChain: B: PDB Molecule: lyr motif-containing protein 4;PDBTitle: crystal structure of human nfs1-isd11 in complex with e. coli acyl-2 carrier protein at 3.09 angstroms
19 c2kjwA_
8.0
16
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s6;PDBTitle: solution structure and backbone dynamics of the permutant p54-55
20 d2jfga2
7.0
7
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
21 c3t0yA_
not modelled
6.9
13
PDB header: transcription regulator/protein bindingChain: A: PDB Molecule: response regulator;PDBTitle: structure of the phyr anti-anti-sigma domain bound to the anti-sigma2 factor, nepr
22 c1tteA_
not modelled
6.7
9
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-conjugating enzyme e2-24 kda;PDBTitle: the structure of a class ii ubiquitin-conjugating enzyme,2 ubc1.
23 c4whnB_
not modelled
6.4
9
PDB header: transferaseChain: B: PDB Molecule: apxc;PDBTitle: structure of toxin-activating acyltransferase (taat)
24 c1t3oA_
not modelled
6.1
19
PDB header: rna binding proteinChain: A: PDB Molecule: carbon storage regulator;PDBTitle: solution structure of csra, a bacterial carbon storage2 regulatory protein
25 d1xxaa_
not modelled
6.1
14
Fold: DCoH-likeSuperfamily: C-terminal domain of arginine repressorFamily: C-terminal domain of arginine repressor
26 d1gzga_
not modelled
5.9
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
27 d2c1ha1
not modelled
5.8
8
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
28 d1l6sa_
not modelled
5.8
8
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
29 d1k8kd1
not modelled
5.8
18
Fold: Secretion chaperone-likeSuperfamily: Arp2/3 complex subunitsFamily: Arp2/3 complex subunits
30 c3obkH_
not modelled
5.7
8
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
31 d1pv8a_
not modelled
5.7
4
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)