Experiment: We designed an M. tuberculosis CRISPRi library (RLC12) to target all annotated Mtb genes with single guide RNAs (sgRNAs) of varying predicted knockdown efficiencies.
In parallel, we constructed a M. smegmatis CRISPRi library (RLC11) to target all annotated Msmeg genes, similar to the Mtb sgRNA library.
RLC12 was transformed into M. tuberculosis H37Rv; RLC11 into M. smegmatis mc2-155.
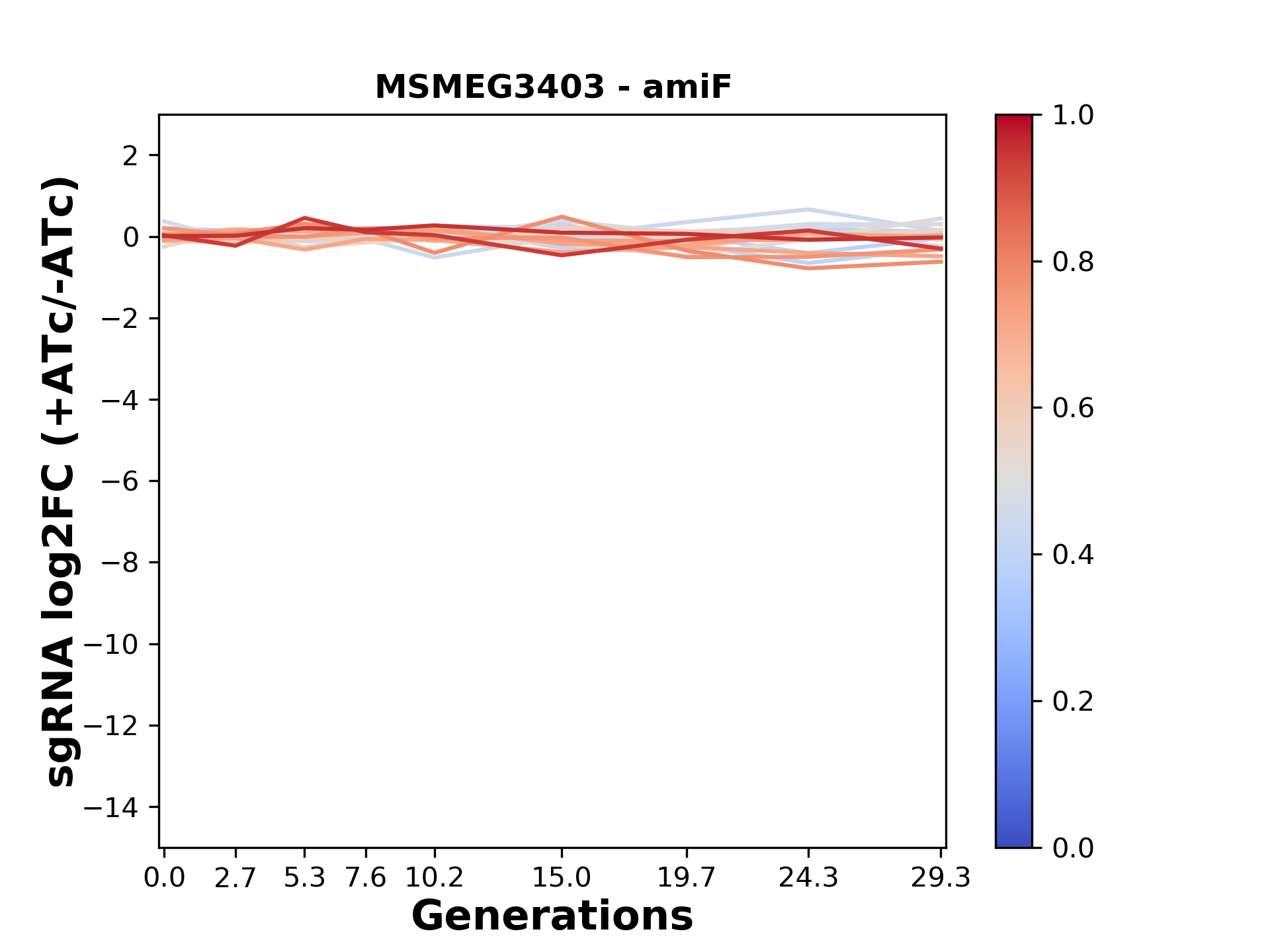
We passaged both CRISPRi libraries for nearly 30 generations in the presence or absence of the CRISPRi-inducer anhydrotetracycline (ATc). At the indicated timepoints, genomic DNA was harvested and the representation of individual sgRNAs was counted using Illumina sequencing.
Here, we show sgRNA log2 fold-change values (L2FC) (plus versus minus ATc) over consecutive generations for sgRNAs targeting the gene you selected.
sgRNAs are color-coded based on their predicted strength from blue (strength=0; weak) to red (strength=1; strong).
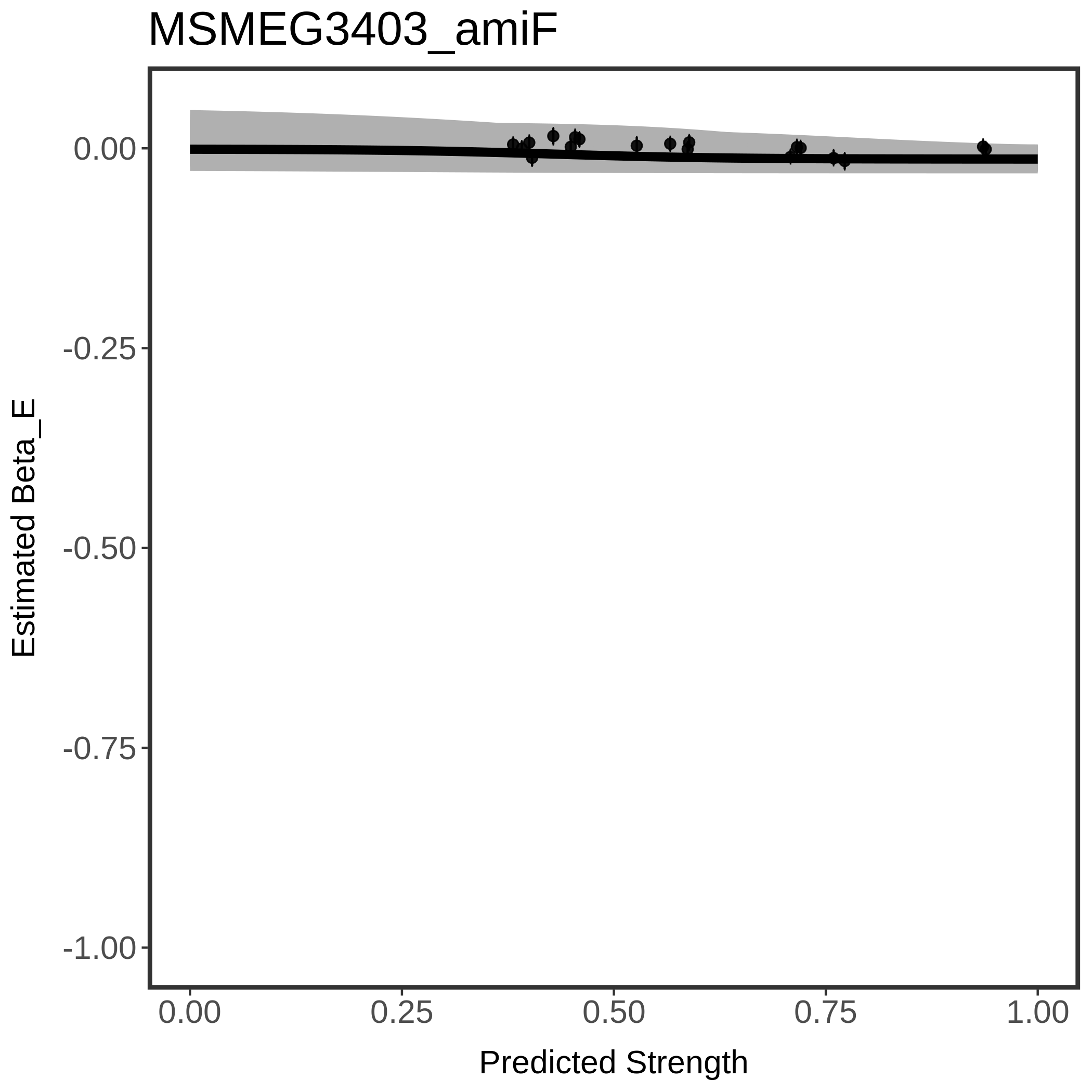
In order to quantify gene vulnerability, data were subsequently analyzed with a hierarchical model at the sgRNA level (two-line model) and gene level (Hill curve). The vulnerability quantification is shown below. [More information]