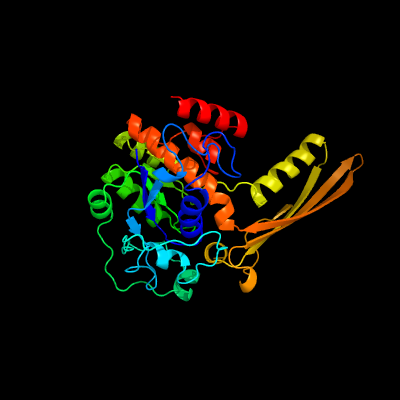
1 c1gr0A_
100.0
100
PDB header: isomeraseChain: A: PDB Molecule: inositol-3-phosphate synthase;PDBTitle: myo-inositol 1-phosphate synthase from mycobacterium2 tuberculosis in complex with nad and zinc.
2 c3cinA_
100.0
29
PDB header: isomeraseChain: A: PDB Molecule: myo-inositol-1-phosphate synthase-related protein;PDBTitle: crystal structure of a myo-inositol-1-phosphate synthase-related2 protein (tm_1419) from thermotoga maritima msb8 at 1.70 a resolution
3 c1u1iC_
100.0
20
PDB header: isomeraseChain: C: PDB Molecule: myo-inositol-1-phosphate synthase;PDBTitle: myo-inositol phosphate synthase mips from a. fulgidus
4 c1vkoA_
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: inositol-3-phosphate synthase;PDBTitle: crystal structure of inositol-3-phosphate synthase (ce21227) from2 caenorhabditis elegans at 2.30 a resolution
5 c1p1hD_
100.0
21
PDB header: isomeraseChain: D: PDB Molecule: inositol-3-phosphate synthase;PDBTitle: crystal structure of the 1l-myo-inositol/nad+ complex
6 d1gr0a1
100.0
100
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
7 d1vjpa1
100.0
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
8 d1u1ia1
100.0
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
9 d1p1ja1
100.0
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
10 d1vkoa1
100.0
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
11 d1vkoa2
100.0
21
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
12 d1u1ia2
100.0
23
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
13 d1p1ja2
100.0
20
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
14 d1vjpa2
100.0
25
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
15 d1gr0a2
99.9
100
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
16 c3upyB_
98.2
18
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of the brucella abortus enzyme catalyzing the first2 committed step of the methylerythritol 4-phosphate pathway.
17 c3wb9A_
98.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: diaminopimelate dehydrogenase;PDBTitle: crystal structures of meso-diaminopimelate dehydrogenase from2 symbiobacterium thermophilum
18 c3mtjA_
98.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
19 c6iaqA_
98.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase n-terminus domain-containingPDBTitle: structure of amine dehydrogenase from mycobacterium smegmatis
20 c6iauB_
98.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: amine dehydrogenase;PDBTitle: amine dehydrogenase from cystobacter fuscus in complex with nadp+ and2 cyclohexylamine
21 c3bioB_
not modelled
98.0
19
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase, gfo/idh/moca family;PDBTitle: crystal structure of oxidoreductase (gfo/idh/moca family member) from2 porphyromonas gingivalis w83
22 c4xb1B_
not modelled
97.8
17
PDB header: oxidoreductaseChain: B: PDB Molecule: 319aa long hypothetical homoserine dehydrogenase;PDBTitle: hyperthermophilic archaeal homoserine dehydrogenase in complex with2 nadph
23 c3wgzB_
not modelled
97.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: meso-diaminopimelate dehydrogenase;PDBTitle: crystal structure of meso-dapdh q154l/t173i/r199m/p248s/h249n/n276s2 mutant with d-leucine of from clostridium tetani e88
24 c6dzsD_
not modelled
97.6
18
PDB header: oxidoreductaseChain: D: PDB Molecule: homoserine dehydrogenase;PDBTitle: mycobacterial homoserine dehydrogenase thra in complex with nadp
25 c3c8mA_
not modelled
97.3
12
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of homoserine dehydrogenase from thermoplasma2 volcanium
26 c5kt0A_
not modelled
97.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
27 c1drwA_
not modelled
97.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
28 c5zz5D_
not modelled
97.2
25
PDB header: gene regulationChain: D: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: redox-sensing transcriptional repressor rex
29 c4pg8B_
not modelled
97.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of s. aureus homoserine dehydrogenase at ph8.5
30 c2ph5A_
not modelled
97.0
15
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
31 c5tenH_
not modelled
97.0
21
PDB header: oxidoreductaseChain: H: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
32 c3do5A_
not modelled
96.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of putative homoserine dehydrogenase (np_069768.1)2 from archaeoglobus fulgidus at 2.20 a resolution
33 c3ceaA_
not modelled
96.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: myo-inositol 2-dehydrogenase;PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
34 c3wg9D_
not modelled
96.9
19
PDB header: transcriptionChain: D: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: crystal structure of rsp, a rex-family repressor
35 c3ketA_
not modelled
96.9
23
PDB header: transcription/dnaChain: A: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
36 c6g1mA_
not modelled
96.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: amine dehydrogenase from petrotoga mobilis; open and closed form
37 c3ingA_
not modelled
96.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of homoserine dehydrogenase (np_394635.1) from2 thermoplasma acidophilum at 1.95 a resolution
38 c2ejwB_
not modelled
96.8
19
PDB header: oxidoreductaseChain: B: PDB Molecule: homoserine dehydrogenase;PDBTitle: homoserine dehydrogenase from thermus thermophilus hb8
39 c1ebuA_
not modelled
96.8
11
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: homoserine dehydrogenase complex with nad analogue and l-2 homoserine
40 c5l78A_
not modelled
96.8
16
PDB header: oxidoreductaseChain: A: PDB Molecule: alpha-aminoadipic semialdehyde synthase, mitochondrial;PDBTitle: crystal structure of human aminoadipate semialdehyde synthase,2 saccharopine dehydrogenase domain (in nad+ bound form)
41 c1e5lA_
not modelled
96.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine reductase;PDBTitle: apo saccharopine reductase from magnaporthe grisea
42 c2axqA_
not modelled
96.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
43 c3wycB_
not modelled
96.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: meso-diaminopimelate d-dehydrogenase;PDBTitle: structure of a meso-diaminopimelate dehydrogenase in complex with nadp
44 c5ugjC_
not modelled
96.4
23
PDB header: oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
45 c5z2fA_
not modelled
96.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
46 c5avoA_
not modelled
96.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of the reduced form of homoserine dehydrogenase from2 sulfolobus tokodaii.
47 c2ixaA_
not modelled
96.3
24
PDB header: hydrolaseChain: A: PDB Molecule: alpha-n-acetylgalactosaminidase;PDBTitle: a-zyme, n-acetylgalactosaminidase
48 d1ydwa1
not modelled
96.2
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
49 c4inaA_
not modelled
96.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: crystal structure of the q7mss8_wolsu protein from wolinella2 succinogenes. northeast structural genomics consortium target wsr35
50 c4gqaC_
not modelled
96.2
20
PDB header: oxidoreductaseChain: C: PDB Molecule: nad binding oxidoreductase;PDBTitle: crystal structure of nad binding oxidoreductase from klebsiella2 pneumoniae
51 d2dt5a2
not modelled
96.1
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Transcriptional repressor Rex, C-terminal domain
52 c2dt5A_
not modelled
96.0
26
PDB header: dna binding proteinChain: A: PDB Molecule: at-rich dna-binding protein;PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
53 c3e18A_
not modelled
96.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of nad-binding protein from listeria innocua
54 d1ebfa1
not modelled
95.6
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
55 c3dapB_
not modelled
95.6
22
PDB header: oxidoreductaseChain: B: PDB Molecule: diaminopimelic acid dehydrogenase;PDBTitle: c. glutamicum dap dehydrogenase in complex with nadp+ and2 the inhibitor 5s-isoxazoline
56 c3kuxA_
not modelled
95.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase;PDBTitle: structure of the ypo2259 putative oxidoreductase from yersinia pestis
57 c2nvwB_
not modelled
95.6
11
PDB header: transcriptionChain: B: PDB Molecule: galactose/lactose metabolism regulatory proteinPDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
58 c5wolA_
not modelled
95.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase dapb from coxiella2 burnetii
59 c4f3yA_
not modelled
95.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: x-ray crystal structure of dihydrodipicolinate reductase from2 burkholderia thailandensis
60 c5yabD_
not modelled
95.3
16
PDB header: oxidoreductaseChain: D: PDB Molecule: scyllo-inositol dehydrogenase with l-glucose dehydrogenasePDBTitle: crystal structure of scyllo-inositol dehydrogenase with l-glucose2 dehydrogenase activity
61 c1j5pA_
not modelled
95.2
24
PDB header: oxidoreductaseChain: A: PDB Molecule: aspartate dehydrogenase;PDBTitle: crystal structure of aspartate dehydrogenase (tm1643) from thermotoga2 maritima at 1.9 a resolution
62 c6a3fB_
not modelled
94.9
10
PDB header: oxidoreductaseChain: B: PDB Molecule: putative dehydrogenase;PDBTitle: levoglucosan dehydrogenase, apo form
63 c3c1aB_
not modelled
94.9
18
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a putative oxidoreductase (zp_00056571.1) from2 magnetospirillum magnetotacticum ms-1 at 1.85 a resolution
64 c5uibA_
not modelled
94.8
21
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase protein;PDBTitle: crystal structure of an oxidoreductase from agrobacterium radiobacter2 in complex with nad+, l-tartaric acid and magnesium
65 c4rl6A_
not modelled
94.8
11
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: crystal structure of the q04l03_strp2 protein from streptococcus2 pneumoniae. northeast structural genomics consortium target spr105
66 d1diha1
not modelled
94.8
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
67 c3ijpA_
not modelled
94.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
68 c4ywjB_
not modelled
94.6
26
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate reductase (htpa2 reductase) from pseudomonas aeruginosa
69 d1f06a1
not modelled
94.6
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
70 c2q4eB_
not modelled
94.6
16
PDB header: oxidoreductaseChain: B: PDB Molecule: probable oxidoreductase at4g09670;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
71 c1lc3A_
not modelled
94.3
20
PDB header: oxidoreductaseChain: A: PDB Molecule: biliverdin reductase a;PDBTitle: crystal structure of a biliverdin reductase enzyme-cofactor2 complex
72 d1j5pa4
not modelled
94.3
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
73 c4mkzA_
not modelled
93.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: inositol dehydrogenase;PDBTitle: crystal structure of apo scyllo-inositol dehydrogenase from2 lactobacillus casei at 77k
74 d1ryda1
not modelled
93.9
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
75 c2dc1A_
not modelled
93.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
76 c2vt2A_
not modelled
93.8
16
PDB header: transcriptionChain: A: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: structure and functional properties of the bacillus subtilis2 transcriptional repressor rex
77 c4bgvB_
not modelled
93.5
17
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: 1.8 a resolution structure of the malate dehydrogenase from2 picrophilus torridus in its apo form
78 d2nvwa1
not modelled
93.3
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
79 c1zh8B_
not modelled
93.3
15
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
80 c4ew6A_
not modelled
92.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: d-galactose-1-dehydrogenase protein;PDBTitle: crystal structure of d-galactose-1-dehydrogenase protein from2 rhizobium etli
81 c4hktA_
not modelled
92.8
22
PDB header: oxidoreductaseChain: A: PDB Molecule: inositol 2-dehydrogenase;PDBTitle: crystal structure of a putative myo-inositol dehydrogenase from2 sinorhizobium meliloti 1021 (target psi-012312)
82 c3q2kB_
not modelled
92.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
83 c4h3vA_
not modelled
92.4
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: oxidoreductase domain protein;PDBTitle: crystal structure of oxidoreductase domain protein from kribbella2 flavida
84 c3nt5B_
not modelled
92.0
13
PDB header: oxidoreductaseChain: B: PDB Molecule: inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
85 c1r0lD_
not modelled
91.9
18
PDB header: oxidoreductaseChain: D: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis2 in complex with nadph
86 c6jnkA_
not modelled
91.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: l-arabinose 1-dehydrogenase (nad(p)(+));PDBTitle: crystal structure of azospirillum brasilense l-arabinose 1-2 dehydrogenase (nadp-bound form)
87 c1llcA_
not modelled
91.7
17
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: structure determination of the allosteric l-lactate dehydrogenase from2 lactobacillus casei at 3.0 angstroms resolution
88 c2z2vA_
not modelled
91.5
17
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein ph1688;PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
89 c3dtyA_
not modelled
91.4
23
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, gfo/idh/moca family;PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
90 c3a14B_
not modelled
91.3
16
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
91 c4hadD_
not modelled
91.1
16
PDB header: oxidoreductaseChain: D: PDB Molecule: probable oxidoreductase protein;PDBTitle: crystal structure of probable oxidoreductase protein from rhizobium2 etli cfn 42
92 c3btuD_
not modelled
90.9
13
PDB header: transcriptionChain: D: PDB Molecule: galactose/lactose metabolism regulatory proteinPDBTitle: crystal structure of the super-repressor mutant of gal80p2 from saccharomyces cerevisiae; gal80(s2) [e351k]
93 d1zh8a1
not modelled
90.9
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
94 c1h6dL_
not modelled
90.5
18
PDB header: protein translocationChain: L: PDB Molecule: precursor form of glucose-fructosePDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
95 c1ofgF_
not modelled
90.5
18
PDB header: oxidoreductaseChain: F: PDB Molecule: glucose-fructose oxidoreductase;PDBTitle: glucose-fructose oxidoreductase
96 c3ezyB_
not modelled
90.1
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: dehydrogenase;PDBTitle: crystal structure of probable dehydrogenase tm_0414 from thermotoga2 maritima
97 c3m2tA_
not modelled
90.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: probable dehydrogenase;PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
98 c5eesA_
not modelled
90.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal strcuture of dapb in complex with nadp+ from corynebacterium2 glutamicum
99 c2glxD_
not modelled
89.8
19
PDB header: oxidoreductaseChain: D: PDB Molecule: 1,5-anhydro-d-fructose reductase;PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
100 d1u8xx1
not modelled
89.4
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
101 c3euwB_
not modelled
89.3
15
PDB header: oxidoreductaseChain: B: PDB Molecule: myo-inositol dehydrogenase;PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
102 c4q3nA_
not modelled
89.2
15
PDB header: hydrolaseChain: A: PDB Molecule: mgs-m5;PDBTitle: crystal structure of mgs-m5, a lactate dehydrogenase enzyme from a2 medee basin deep-sea metagenome library
103 c3ic5A_
not modelled
88.6
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
104 c6norB_
not modelled
88.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nad dependent dehydrogenase;PDBTitle: crystal structure of gend2 from gentamicin a biosynthesis in complex2 with nad
105 c3d0oA_
not modelled
88.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase 1;PDBTitle: crystal structure of lactate dehydrogenase from staphylococcus aureus
106 c4gmfD_
not modelled
88.1
14
PDB header: oxidoreductaseChain: D: PDB Molecule: yersiniabactin biosynthetic protein ybtu;PDBTitle: apo structure of a thiazolinyl imine reductase from yersinia2 enterocolitica (irp3)
107 d1yl7a1
not modelled
88.1
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
108 d5ldha1
not modelled
87.8
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
109 c1ez4B_
not modelled
87.7
16
PDB header: oxidoreductaseChain: B: PDB Molecule: lactate dehydrogenase;PDBTitle: crystal structure of non-allosteric l-lactate dehydrogenase2 from lactobacillus pentosus at 2.3 angstrom resolution
110 c3gfgB_
not modelled
87.4
15
PDB header: oxidoreductaseChain: B: PDB Molecule: uncharacterized oxidoreductase yvaa;PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
111 c3evnA_
not modelled
87.2
14
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, gfo/idh/moca family;PDBTitle: crystal structure of putative oxidoreductase from streptococcus2 agalactiae 2603v/r
112 c5b3uB_
not modelled
87.0
16
PDB header: transferaseChain: B: PDB Molecule: biliverdin reductase;PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
113 c3tl2A_
not modelled
86.2
13
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of bacillus anthracis str. ames malate dehydrogenase2 in closed conformation.
114 c2fnzA_
not modelled
86.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: lactate dehydrogenase;PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
115 c3moiA_
not modelled
85.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: probable dehydrogenase;PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
116 d2g0ta1
not modelled
85.1
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
117 c2ldxA_
not modelled
84.9
17
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: apo-lactate dehydrogenase;PDBTitle: characterization of the antigenic sites on the refined 3-2 angstroms resolution structure of mouse testicular lactate3 dehydrogenase c4
118 c2ozpA_
not modelled
84.5
21
PDB header: oxidoreductaseChain: A: PDB Molecule: n-acetyl-gamma-glutamyl-phosphate reductase;PDBTitle: crystal structure of n-acetyl-gamma-glutamyl-phosphate reductase2 (ttha1904) from thermus thermophilus
119 c3rbvA_
not modelled
84.3
10
PDB header: sugar binding proteinChain: A: PDB Molecule: sugar 3-ketoreductase;PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
120 c8ldhA_
not modelled
83.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: m4 apo-lactate dehydrogenase;PDBTitle: refined crystal structure of dogfish m4 apo-lactate dehydrogenase