1 c2we7A_
100.0
29
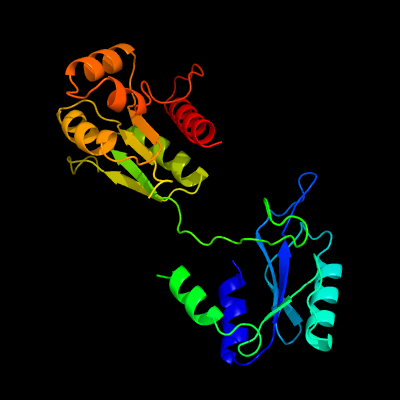
PDB header: oxidoreductaseChain: A: PDB Molecule: xanthine dehydrogenase;PDBTitle: crystal structure of mycobacterium tuberculosis rv0376c2 homologue from mycobacterium smegmatis
2 c3on5B_
100.0
23
PDB header: oxidoreductaseChain: B: PDB Molecule: bh1974 protein;PDBTitle: crystal structure of a xanthine dehydrogenase (bh1974) from bacillus2 halodurans at 2.80 a resolution
3 c3llvA_
97.5
14
PDB header: nad(p) binding proteinChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
4 c2g1uA_
97.3
14
PDB header: transport proteinChain: A: PDB Molecule: hypothetical protein tm1088a;PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
5 c5nc8B_
96.8
16
PDB header: transport proteinChain: B: PDB Molecule: potassium efflux system protein;PDBTitle: shewanella denitrificans kef ctd in amp bound form
6 d1lssa_
96.5
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
7 c4g65A_
96.5
12
PDB header: transport proteinChain: A: PDB Molecule: trk system potassium uptake protein trka;PDBTitle: potassium transporter peripheral membrane component (trka) from vibrio2 vulnificus
8 c3ic5A_
96.4
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
9 c1dxyA_
96.4
25
PDB header: oxidoreductaseChain: A: PDB Molecule: d-2-hydroxyisocaproate dehydrogenase;PDBTitle: structure of d-2-hydroxyisocaproate dehydrogenase
10 c4gx5D_
96.3
11
PDB header: transport proteinChain: D: PDB Molecule: trka domain protein;PDBTitle: gsuk channel
11 c3fwzA_
96.3
12
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein ybal;PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
12 d1kjqa2
96.3
17
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
13 d2hmva1
96.1
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
14 c2cukC_
96.1
19
PDB header: oxidoreductaseChain: C: PDB Molecule: glycerate dehydrogenase/glyoxylate reductase;PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
15 c3q2oB_
96.1
14
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
16 c2dbqA_
96.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glyoxylate reductase;PDBTitle: crystal structure of glyoxylate reductase (ph0597) from pyrococcus2 horikoshii ot3, complexed with nadp (i41)
17 c3l4bG_
96.1
18
PDB header: transport proteinChain: G: PDB Molecule: trka k+ channel protien tm1088b;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
18 c3evtA_
96.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: phosphoglycerate dehydrogenase;PDBTitle: crystal structure of phosphoglycerate dehydrogenase from lactobacillus2 plantarum
19 c1j4aA_
95.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: insights into domain closure, substrate specificity and2 catalysis of d-lactate dehydrogenase from lactobacillus3 bulgaricus
20 c6ih2B_
95.9
19
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphite dehydrogenase;PDBTitle: crystal structure of phosphite dehydrogenase from ralstonia sp. 4506
21 c5j23D_
not modelled
95.9
21
PDB header: oxidoreductaseChain: D: PDB Molecule: 2-hydroxyacid dehydrogenase;PDBTitle: crystal structure of nadph-dependent glyoxylate/hydroxypyruvate2 reductase smc04462 (smghrb) from sinorhizobium meliloti in complex3 with 2'-phospho-adp-ribose
22 d2jfga1
not modelled
95.9
20
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
23 c5tulA_
not modelled
95.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: tetracycline destructase tet(55);PDBTitle: crystal structure of tetracycline destructase tet(55)
24 c4zqbB_
not modelled
95.9
33
PDB header: oxidoreductaseChain: B: PDB Molecule: nadp-dependent dehydrogenase;PDBTitle: crystal structure of nadp-dependent dehydrogenase from2 rhodobactersphaeroides in complex with nadp and sulfate
25 c4g2nA_
not modelled
95.8
24
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase, nad-binding;PDBTitle: crystal structure of putative d-isomer specific 2-hydroxyacid2 dehydrogenase, nad-binding from polaromonas sp. js6 66
26 c5tx7A_
not modelled
95.8
28
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase familyPDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from desulfovibrio vulgaris
27 d1dxya1
not modelled
95.8
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
28 c3wwyA_
not modelled
95.8
27
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: the crystal structure of d-lactate dehydrogenase from fusobacterium2 nucleatum subsp. nucleatum
29 c3eywA_
not modelled
95.8
14
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
30 d3c96a1
not modelled
95.7
16
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
31 c3k5pA_
not modelled
95.7
31
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
32 c3uvzB_
not modelled
95.7
12
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
33 c3bazA_
not modelled
95.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxyphenylpyruvate reductase;PDBTitle: structure of hydroxyphenylpyruvate reductase from coleus blumei in2 complex with nadp+
34 c2d0iC_
not modelled
95.7
18
PDB header: oxidoreductaseChain: C: PDB Molecule: dehydrogenase;PDBTitle: crystal structure ph0520 protein from pyrococcus horikoshii ot3
35 c3kboB_
not modelled
95.7
24
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxylate/hydroxypyruvate reductase a;PDBTitle: 2.14 angstrom crystal structure of putative oxidoreductase (ycdw) from2 salmonella typhimurium in complex with nadp
36 c1wwkA_
not modelled
95.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: phosphoglycerate dehydrogenase;PDBTitle: crystal structure of phosphoglycerate dehydrogenase from pyrococcus2 horikoshii ot3
37 c1ybaC_
not modelled
95.6
30
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: the active form of phosphoglycerate dehydrogenase
38 c3gmbB_
not modelled
95.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase
39 c2eklA_
not modelled
95.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: structure of st1218 protein from sulfolobus tokodaii
40 d1id1a_
not modelled
95.6
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain
41 c4weqA_
not modelled
95.5
30
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent dehydrogenase;PDBTitle: crystal structure of nadph-dependent glyoxylate/hydroxypyruvate2 reductase smc02828 (smghra) from sinorhizobium meliloti in complex3 with nadp and sulfate
42 c3allA_
not modelled
95.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
43 c4prkB_
not modelled
95.5
19
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-phosphoerythronate dehydrogenase;PDBTitle: crystal structure of d-lactate dehydrogenase (d-ldh) from2 lactobacillus jensenii
44 c3orqA_
not modelled
95.5
10
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
45 c4lswA_
not modelled
95.5
15
PDB header: hydrolaseChain: A: PDB Molecule: d-2-hydroxyacid dehydrogensase protein;PDBTitle: crystallization and structural analysis of 2-hydroxyacid dehydrogenase2 from ketogulonicigenium vulgare y25
46 c2gcgB_
not modelled
95.4
21
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxylate reductase/hydroxypyruvate reductase;PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
47 c5mh5A_
not modelled
95.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: d-2-hydroxyacid dehydrogenase;PDBTitle: d-2-hydroxyacid dehydrogenases (d2-hdh) from haloferax mediterranei in2 complex with 2-keto-hexanoic acid and nadp+ (1.4 a resolution)
48 c3wwzB_
not modelled
95.4
22
PDB header: oxidoreductaseChain: B: PDB Molecule: d-lactate dehydrogenase (fermentative);PDBTitle: the crystal structure of d-lactate dehydrogenase from pseudomonas2 aeruginosa
49 d1mx3a1
not modelled
95.4
27
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
50 c4txkA_
not modelled
95.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: protein-methionine sulfoxide oxidase mical1;PDBTitle: construct of mical-1 containing the monooxygenase and calponin2 homology domains
51 d1ygya1
not modelled
95.3
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
52 c5fn0C_
not modelled
95.3
15
PDB header: oxidoreductaseChain: C: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of pseudomonas fluorescens kynurenine-3-2 monooxygenase (kmo) in complex with gsk180
53 d1j4aa1
not modelled
95.3
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
54 c4n18A_
not modelled
95.3
36
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase familyPDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 family protein from klebsiella pneumoniae 342
55 c2omeA_
not modelled
95.3
26
PDB header: oxidoreductaseChain: A: PDB Molecule: c-terminal-binding protein 2;PDBTitle: crystal structure of human ctbp2 dehydrogenase complexed with nad(h)
56 d2dlda1
not modelled
95.3
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
57 c2g76A_
not modelled
95.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
58 c4cukA_
not modelled
95.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: structure of salmonella d-lactate dehydrogenase in complex2 with nadh
59 d1sc6a1
not modelled
95.3
27
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
60 c2pi1C_
not modelled
95.2
15
PDB header: oxidoreductaseChain: C: PDB Molecule: d-lactate dehydrogenase;PDBTitle: crystal structure of d-lactate dehydrogenase from aquifex2 aeolicus complexed with nad and lactic acid
61 c4gvlB_
not modelled
95.2
10
PDB header: transport proteinChain: B: PDB Molecule: trka domain protein;PDBTitle: crystal structure of the gsuk rck domain
62 c4njmA_
not modelled
95.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase, putative;PDBTitle: crystal structure of phosphoglycerate bound 3-phosphoglycerate2 dehydrogenase in entamoeba histolytica
63 c4gx2B_
not modelled
95.2
11
PDB header: transport proteinChain: B: PDB Molecule: trka domain protein;PDBTitle: gsuk channel bound to nad
64 c3n7uD_
not modelled
95.2
22
PDB header: oxidoreductaseChain: D: PDB Molecule: formate dehydrogenase;PDBTitle: nad-dependent formate dehydrogenase from higher-plant arabidopsis2 thaliana in complex with nad and azide
65 c2yq4C_
not modelled
95.1
19
PDB header: oxidoreductaseChain: C: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase;PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from lactobacillus delbrueckii ssp. bulgaricus
66 c3gvxA_
not modelled
95.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerate dehydrogenase related protein;PDBTitle: crystal structure of glycerate dehydrogenase related2 protein from thermoplasma acidophilum
67 c2qa2A_
not modelled
95.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: polyketide oxygenase cabe;PDBTitle: crystal structure of cabe, an aromatic hydroxylase from angucycline2 biosynthesis, determined to 2.7 a resolution
68 c2xdoC_
not modelled
95.1
20
PDB header: oxidoreductaseChain: C: PDB Molecule: tetx2 protein;PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
69 c1gdhA_
not modelled
95.0
25
PDB header: oxidoreductase(choh (d)-nad(p)+ (a))Chain: A: PDB Molecule: d-glycerate dehydrogenase;PDBTitle: crystal structure of a nad-dependent d-glycerate2 dehydrogenase at 2.4 angstroms resolution
70 c4bk2A_
not modelled
95.0
11
PDB header: oxidoreductaseChain: A: PDB Molecule: probable salicylate monooxygenase;PDBTitle: crystal structure of 3-hydroxybenzoate 6-hydroxylase2 uncovers lipid-assisted flavoprotein strategy for3 regioselective aromatic hydroxylation: q301e mutant
71 c5eowA_
not modelled
95.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-hydroxynicotinate 3-monooxygenase;PDBTitle: crystal structure of 6-hydroxynicotinic acid 3-monooxygenase from2 pseudomonas putida kt2440
72 c5tukC_
not modelled
94.9
16
PDB header: oxidoreductaseChain: C: PDB Molecule: tetracycline destructase tet(51);PDBTitle: crystal structure of tetracycline destructase tet(51)
73 c4xkjA_
not modelled
94.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: a novel d-lactate dehydrogenase from sporolactobacillus sp
74 c5xgvB_
not modelled
94.9
20
PDB header: oxidoreductaseChain: B: PDB Molecule: pyre3;PDBTitle: the structure of diels-alderase pyre3 in the biosynthetic pathway of2 pyrroindomycins
75 d1qp8a1
not modelled
94.9
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
76 c4xcvA_
not modelled
94.9
23
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp-dependent 2-hydroxyacid dehydrogenase;PDBTitle: probable 2-hydroxyacid dehydrogenase from rhizobium etli cfn 42 in2 complex with nadph
77 c2vouA_
not modelled
94.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: 2,6-dihydroxypyridine hydroxylase;PDBTitle: structure of 2,6-dihydroxypyridine-3-hydroxylase from2 arthrobacter nicotinovorans
78 c4cy8A_
not modelled
94.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-hydroxybiphenyl 3-monooxygenase;PDBTitle: 2-hydroxybiphenyl 3-monooxygenase (hbpa) in complex with fad
79 c4hy3D_
not modelled
94.8
20
PDB header: oxidoreductaseChain: D: PDB Molecule: phosphoglycerate oxidoreductase;PDBTitle: crystal structure of a phosphoglycerate oxidoreductase from rhizobium2 etli
80 c2qa1A_
not modelled
94.8
10
PDB header: oxidoreductaseChain: A: PDB Molecule: polyketide oxygenase pgae;PDBTitle: crystal structure of pgae, an aromatic hydroxylase involved in2 angucycline biosynthesis
81 c1qp8A_
not modelled
94.7
12
PDB header: oxidoreductaseChain: A: PDB Molecule: formate dehydrogenase;PDBTitle: crystal structure of a putative formate dehydrogenase from2 pyrobaculum aerophilum
82 c1xdwA_
not modelled
94.7
15
PDB header: oxidoreductaseChain: A: PDB Molecule: nad+-dependent (r)-2-hydroxyglutaratePDBTitle: nad+-dependent (r)-2-hydroxyglutarate dehydrogenase from2 acidaminococcus fermentans
83 c5butG_
not modelled
94.7
9
PDB header: membrane proteinChain: G: PDB Molecule: ktr system potassium uptake protein a,ktr system potassiumPDBTitle: crystal structure of inactive conformation of ktrab k+ transporter
84 c1ygyA_
not modelled
94.7
30
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
85 d1e5qa1
not modelled
94.7
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
86 d2voua1
not modelled
94.7
11
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
87 c3hg7A_
not modelled
94.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase familyPDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 family protein from aeromonas salmonicida subsp. salmonicida a449
88 c4j7cA_
not modelled
94.6
7
PDB header: transport proteinChain: A: PDB Molecule: ktr system potassium uptake protein a;PDBTitle: ktrab potassium transporter from bacillus subtilis
89 c5tueB_
not modelled
94.6
16
PDB header: oxidoreductaseChain: B: PDB Molecule: tetracycline destructase tet(50);PDBTitle: crystal structure of tetracycline destructase tet(50)
90 c4j33B_
not modelled
94.6
14
PDB header: oxidoreductaseChain: B: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of kynurenine 3-monooxygenase (kmo-394)
91 c3rp7A_
not modelled
94.6
8
PDB header: oxidoreductaseChain: A: PDB Molecule: flavoprotein monooxygenase;PDBTitle: crystal structure of klebsiella pneumoniae hpxo complexed with fad and2 uric acid
92 c4e5kC_
not modelled
94.6
18
PDB header: oxidoreductaseChain: C: PDB Molecule: phosphite dehydrogenase (thermostable variant);PDBTitle: thermostable phosphite dehydrogenase in complex with nad and sulfite
93 c2rgjA_
not modelled
94.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: flavin-containing monooxygenase;PDBTitle: crystal structure of flavin-containing monooxygenase phzs
94 c4xa8A_
not modelled
94.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase nad-binding;PDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from xanthobacter autotrophicus py2
95 c5kowA_
not modelled
94.5
14
PDB header: oxidoreductaseChain: A: PDB Molecule: pentachlorophenol 4-monooxygenase;PDBTitle: structure of rifampicin monooxygenase
96 c2w2kB_
not modelled
94.5
22
PDB header: oxidoreductaseChain: B: PDB Molecule: d-mandelate dehydrogenase;PDBTitle: crystal structure of the apo forms of rhodotorula graminis2 d-mandelate dehydrogenase at 1.8a.
97 c2j6iC_
not modelled
94.5
25
PDB header: oxidoreductaseChain: C: PDB Molecule: formate dehydrogenase;PDBTitle: candida boidinii formate dehydrogenase (fdh) c-terminal mutant
98 c3k5iB_
not modelled
94.5
12
PDB header: lyaseChain: B: PDB Molecule: phosphoribosyl-aminoimidazole carboxylase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthase from2 aspergillus clavatus in complex with adp and 5-aminoimadazole3 ribonucleotide
99 c4egjD_
not modelled
94.4
24
PDB header: ligaseChain: D: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of d-alanine-d-alanine ligase from burkholderia2 xenovorans
100 c2bryA_
not modelled
94.4
13
PDB header: transportChain: A: PDB Molecule: nedd9 interacting protein with calponin homologyPDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
101 d2naca1
not modelled
94.4
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
102 c2r0gB_
not modelled
94.4
19
PDB header: oxidoreductaseChain: B: PDB Molecule: rebc;PDBTitle: chromopyrrolic acid-soaked rebc with bound 7-carboxy-k252c
103 c4hb9A_
not modelled
94.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: similarities with probable monooxygenase;PDBTitle: crystal structure of a putative fad containing monooxygenase from2 photorhabdus luminescens subsp. laumondii tto1 (target psi-012791)
104 c3c4aA_
not modelled
94.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: probable tryptophan hydroxylase viod;PDBTitle: crystal structure of viod hydroxylase in complex with fad from2 chromobacterium violaceum. northeast structural genomics consortium3 target cvr158
105 d1gdha1
not modelled
94.2
31
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
106 d1reoa1
not modelled
94.2
5
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
107 c2dc1A_
not modelled
94.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
108 c2nacA_
not modelled
94.2
21
PDB header: oxidoreductase(aldehyde(d),nad+(a))Chain: A: PDB Molecule: nad-dependent formate dehydrogenase;PDBTitle: high resolution structures of holo and apo formate dehydrogenase
109 c5x68B_
not modelled
94.2
8
PDB header: oxidoreductaseChain: B: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of human kmo
110 c3fn4A_
not modelled
94.1
25
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent formate dehydrogenase;PDBTitle: apo-form of nad-dependent formate dehydrogenase from bacterium2 moraxella sp.c-1 in closed conformation
111 c3wnvA_
not modelled
93.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: glyoxylate reductase;PDBTitle: crystal structure of a glyoxylate reductase from paecilomyes2 thermophila
112 c2e1mA_
not modelled
93.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: l-glutamate oxidase;PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
113 c1kjjA_
not modelled
93.9
16
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase 2;PDBTitle: crystal structure of glycniamide ribonucleotide transformylase in2 complex with mg-atp-gamma-s
114 c3fmwC_
not modelled
93.9
14
PDB header: oxidoreductaseChain: C: PDB Molecule: oxygenase;PDBTitle: the crystal structure of mtmoiv, a baeyer-villiger monooxygenase from2 the mithramycin biosynthetic pathway in streptomyces argillaceus.
115 d1ps9a3
not modelled
93.9
11
Fold: Nucleotide-binding domainSuperfamily: Nucleotide-binding domainFamily: N-terminal domain of adrenodoxin reductase-like
116 c3ax6C_
not modelled
93.9
15
PDB header: ligaseChain: C: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide2 synthetase from thermotoga maritima
117 c4k2xB_
not modelled
93.9
19
PDB header: oxidoreductase, flavoproteinChain: B: PDB Molecule: polyketide oxygenase/hydroxylase;PDBTitle: oxys anhydrotetracycline hydroxylase from streptomyces rimosus
118 d1l7da1
not modelled
93.8
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
119 c6j0zC_
not modelled
93.7
16
PDB header: oxidoreductaseChain: C: PDB Molecule: putative angucycline-like polyketide oxygenase;PDBTitle: crystal structure of alpk
120 c4mamB_
not modelled
93.7
15
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: the crystal structure of phosphoribosylaminoimidazole carboxylase2 atpase subunit of francisella tularensis subsp. tularensis schu s4 in3 complex with an adp analog, amp-cp