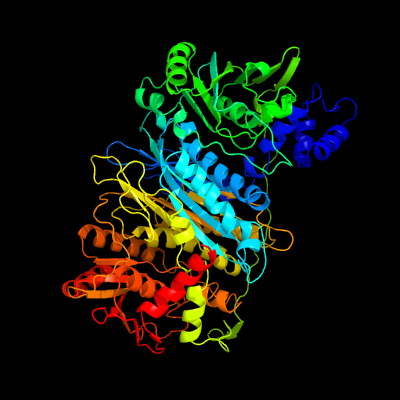
| 1 | c1t3tA_
|
|
 |
100.0 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase;
PDBTitle: structure of formylglycinamide synthetase
|
|
|
|
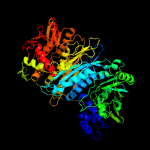
| 2 | c3ac6A_
|
|
 |
100.0 |
43 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase 2;
PDBTitle: crystal structure of purl from thermus thermophilus
|
|
|
|
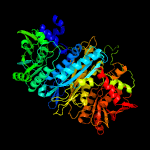
| 3 | c3d54I_
|
|
 |
100.0 |
39 |
PDB header:ligase
Chain: I: PDB Molecule:phosphoribosylformylglycinamidine synthase ii;
PDBTitle: structure of purlqs from thermotoga maritima
|
|
|
|
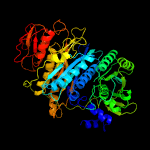
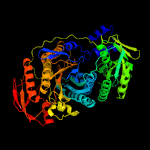
| 4 | c2hs0A_
|
|
 |
100.0 |
40 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase ii;
PDBTitle: t. maritima purl complexed with atp
|
|
|
|
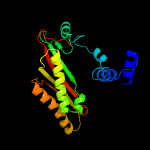
| 5 | d1vk3a1
|
|
 |
100.0 |
51 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
|
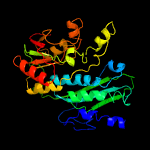
| 6 | c2btuB_
|
|
 |
100.0 |
19 |
PDB header:synthase
Chain: B: PDB Molecule:phosphoribosyl-aminoimidazole synthetase;
PDBTitle: crystal structure of phosphoribosylformylglycinamidine cyclo-ligase2 from bacillus anthracis at 2.3a resolution.
|
|
|
|
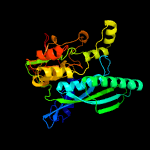
| 7 | c3vysC_
|
|
 |
100.0 |
20 |
PDB header:metal binding protein/transferase
Chain: C: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of the hypc-hypd-hype complex (form i)
|
|
|
|
| 8 | c3mcqA_
|
|
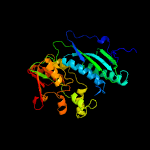 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine-monophosphate kinase;
PDBTitle: crystal structure of thiamine-monophosphate kinase (mfla_0573) from2 methylobacillus flagellatus kt at 1.91 a resolution
|
|
|
|
| 9 | c5cm7A_
|
|
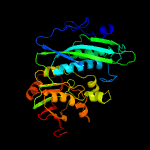 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine-monophosphate kinase;
PDBTitle: structure of thiamine-monophosphate kinase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp) and thiamine3 diphosphate (tpp)
|
|
|
|
| 10 | c5vk4B_
|
|
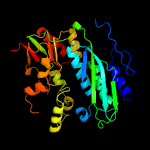 |
100.0 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a phosphoribosylformylglycinamidine cyclo-ligase2 from neisseria gonorrhoeae bound to amppnp and magnesium
|
|
|
|
| 11 | c3fd5B_
|
|
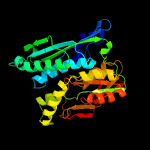 |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase 1;
PDBTitle: crystal structure of human selenophosphate synthetase 12 complex with ampcp
|
|
|
|
| 12 | c1cliD_
|
|
 |
100.0 |
18 |
PDB header:ligase
Chain: D: PDB Molecule:protein (phosphoribosyl-aminoimidazole synthetase);
PDBTitle: x-ray crystal structure of aminoimidazole ribonucleotide synthetase2 (purm), from the e. coli purine biosynthetic pathway, at 2.5 a3 resolution
|
|
|
|
| 13 | c3u0oA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:selenide, water dikinase;
PDBTitle: the crystal structure of selenophosphate synthetase from e. coli
|
|
|
|
| 14 | c2z1eA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hype from thermococcus kodakaraensis (outward2 form)
|
|
|
|
| 15 | c2zodB_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of selenophosphate synthetase from aquifex aeolicus
|
|
|
|
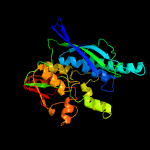
| 16 | c2zauB_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of an n-terminally truncated2 selenophosphate synthetase from aquifex aeolicus
|
|
|
|
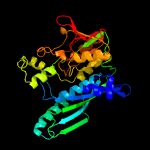
| 17 | c3vtiD_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:hydrogenase maturation factor;
PDBTitle: crystal structure of hype-hypf complex
|
|
|
|
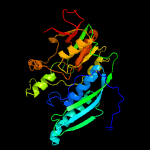
| 18 | c2z1tA_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hydrogenase maturation protein hype
|
|
|
|
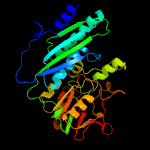
| 19 | c2rb9D_
|
|
 |
100.0 |
18 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hype protein;
PDBTitle: crystal structure of e.coli hype
|
|
|
|
| 20 | d1t3ta5
|
|
 |
100.0 |
22 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
|
| 21 | c3m84A_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 francisella tularensis
|
|
|
| 22 | c2yxzA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin-monophosphate kinase;
PDBTitle: crystal structure of tt0281 from thermus thermophilus hb8
|
|
|
| 23 | c2z01A_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 geobacillus kaustophilus
|
|
|
| 24 | c3c9uB_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:thiamine monophosphate kinase;
PDBTitle: aathil complexed with adp and tpp
|
|
|
| 25 | c2v9yA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: human aminoimidazole ribonucleotide synthetase
|
|
|
| 26 | d1vk3a2 |
|
not modelled |
100.0 |
32 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 27 | c5l16A_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:putative selenophosphate synthetase;
PDBTitle: crystal structure of n-terminus truncated selenophosphate synthetase2 from leishmania major
|
|
|
| 28 | c1vqvB_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:thiamine monophosphate kinase;
PDBTitle: crystal structure of thiamine monophosphate kinase (thil)2 from aquifex aeolicus
|
|
|
| 29 | c5avmE_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: E: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structures of 5-aminoimidazole ribonucleotide (air)2 synthetase, purm, from thermus thermophilus
|
|
|
| 30 | d1t3ta6 |
|
not modelled |
100.0 |
31 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 31 | d1t3ta4 |
|
not modelled |
100.0 |
29 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 32 | d1vk3a3 |
|
not modelled |
100.0 |
44 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 33 | c3mdoB_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:putative phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a putative phosphoribosylformylglycinamidine2 cyclo-ligase (bdi_2101) from parabacteroides distasonis atcc 8503 at3 1.91 a resolution
|
|
|
| 34 | d1t3ta7 |
|
not modelled |
100.0 |
24 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 35 | c3kizA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of putative phosphoribosylformylglycinamidine cyclo-2 ligase (yp_676759.1) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
|
|
|
| 36 | d2z1ea2 |
|
not modelled |
99.9 |
19 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 37 | d2zoda2 |
|
not modelled |
99.9 |
16 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 38 | d1clia2 |
|
not modelled |
99.9 |
19 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 39 | d1clia1 |
|
not modelled |
99.9 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 40 | d1clib1 |
|
not modelled |
99.9 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 41 | d3c9ua2 |
|
not modelled |
99.9 |
17 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 42 | d2zaua1 |
|
not modelled |
99.8 |
23 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 43 | d2zoda1 |
|
not modelled |
99.8 |
23 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 44 | d3c9ua1 |
|
not modelled |
99.8 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 45 | d1t3ta1 |
|
not modelled |
99.7 |
32 |
Fold:RuvA C-terminal domain-like
Superfamily:FGAM synthase PurL, linker domain
Family:FGAM synthase PurL, linker domain |
|
|
| 46 | d2z1ea1 |
|
not modelled |
99.7 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 47 | c5kdmD_ |
|
not modelled |
96.9 |
16 |
PDB header:chaperone / dna binding protein
Chain: D: PDB Molecule:major tegument protein;
PDBTitle: crystal structure of ebv tegument protein bnrf1 in complex with2 histone chaperone daxx and histones h3.3-h4
|
|
|
| 48 | d3dhxa1 |
|
not modelled |
60.1 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
|
|
| 49 | c4zc0A_ |
|
not modelled |
53.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: structure of a dodecameric bacterial helicase
|
|
|
| 50 | d1rp3a1 |
|
not modelled |
52.5 |
44 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
|
|
| 51 | d1ku2a1 |
|
not modelled |
50.2 |
47 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
|
|
| 52 | c1l0oC_ |
|
not modelled |
49.7 |
39 |
PDB header:protein binding
Chain: C: PDB Molecule:sigma factor;
PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
|
|
|
| 53 | d1l0oc_ |
|
not modelled |
49.7 |
39 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 54 | c2zc2A_ |
|
not modelled |
46.5 |
21 |
PDB header:replication
Chain: A: PDB Molecule:dnad-like replication protein;
PDBTitle: crystal structure of dnad-like replication protein from streptococcus2 mutans ua159, gi 24377835, residues 127-199
|
|
|
| 55 | d1to3a_ |
|
not modelled |
46.2 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 56 | c3jrkG_ |
|
not modelled |
44.6 |
20 |
PDB header:lyase
Chain: G: PDB Molecule:tagatose 1,6-diphosphate aldolase 2;
PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
|
|
|
| 57 | d1llua2 |
|
not modelled |
39.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
|
|
| 58 | c1yvlB_ |
|
not modelled |
39.0 |
18 |
PDB header:signaling protein
Chain: B: PDB Molecule:signal transducer and activator of transcription
PDBTitle: structure of unphosphorylated stat1
|
|
|
| 59 | d2csua1 |
|
not modelled |
38.7 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 60 | d2qrra1 |
|
not modelled |
38.1 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
|
|
| 61 | c1szpC_ |
|
not modelled |
35.7 |
16 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna repair protein rad51;
PDBTitle: a crystal structure of the rad51 filament
|
|
|
| 62 | c3bkhA_ |
|
not modelled |
35.2 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:lytic transglycosylase;
PDBTitle: crystal structure of the bacteriophage phikz lytic2 transglycosylase, gp144
|
|
|
| 63 | d1pyta_ |
|
not modelled |
34.3 |
8 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 64 | c4nmnA_ |
|
not modelled |
33.7 |
18 |
PDB header:replication
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: aquifex aeolicus replicative helicase (dnab) complexed with adp, at2 3.3 resolution
|
|
|
| 65 | d1o6xa_ |
|
not modelled |
33.7 |
16 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 66 | d1pcaa1 |
|
not modelled |
33.4 |
12 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 67 | d1ayea2 |
|
not modelled |
32.7 |
16 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 68 | d1kwma2 |
|
not modelled |
32.2 |
16 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 69 | d2boaa2 |
|
not modelled |
31.8 |
20 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 70 | d1rjwa2 |
|
not modelled |
30.5 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
|
|
| 71 | d1uufa2 |
|
not modelled |
30.4 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
|
|
| 72 | d1piwa2 |
|
not modelled |
30.4 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
|
|
| 73 | c4y5wD_ |
|
not modelled |
30.2 |
15 |
PDB header:transcription/dna
Chain: D: PDB Molecule:signal transducer and activator of transcription 6;
PDBTitle: transcription factor-dna complex
|
|
|
| 74 | c5l37E_ |
|
not modelled |
29.8 |
16 |
PDB header:structural protein
Chain: E: PDB Molecule:msm0273;
PDBTitle: the structure of the pentameric shell protein msm0273 from the rmm2 microcompartment
|
|
|
| 75 | d1nsaa2 |
|
not modelled |
29.6 |
16 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 76 | c5vt9A_ |
|
not modelled |
29.3 |
11 |
PDB header:motor protein
Chain: A: PDB Molecule:myosin light chain tgmlc1;
PDBTitle: myosin light chain 1 and myoa complex
|
|
|
| 77 | d1pbaa_ |
|
not modelled |
29.0 |
16 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 78 | c4rgwB_ |
|
not modelled |
26.7 |
15 |
PDB header:transferase/transcription
Chain: B: PDB Molecule:transcription initiation factor tfiid subunit 7;
PDBTitle: crystal structure of a taf1-taf7 complex in human transcription factor2 iid
|
|
|
| 79 | d1jqga2 |
|
not modelled |
26.6 |
12 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 80 | c1uusA_ |
|
not modelled |
24.3 |
11 |
PDB header:signal transduction
Chain: A: PDB Molecule:stat protein;
PDBTitle: structure of an activated dictyostelium stat in its2 dna-unbound form
|
|
|
| 81 | c3bgwD_ |
|
not modelled |
24.1 |
18 |
PDB header:replication
Chain: D: PDB Molecule:dnab-like replicative helicase;
PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
|
|
|
| 82 | d2rcfa1 |
|
not modelled |
22.3 |
14 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
|
|
| 83 | c5h29A_ |
|
not modelled |
22.1 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase/glutathione-related protein;
PDBTitle: crystal structure of the ntd_n/c domain of alkylhydroperoxide2 reductase ahpf from enterococcus faecalis (v583)
|
|
|
| 84 | d2pifa1 |
|
not modelled |
21.7 |
19 |
Fold:PSTPO5379-like
Superfamily:PSTPO5379-like
Family:PSTPO5379-like |
|
|
| 85 | d2qswa1 |
|
not modelled |
20.1 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
|
|
| 86 | c2q6tB_ |
|
not modelled |
19.7 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:dnab replication fork helicase;
PDBTitle: crystal structure of the thermus aquaticus dnab monomer
|
|
|
| 87 | d2guka1 |
|
not modelled |
19.7 |
7 |
Fold:PG1857-like
Superfamily:PG1857-like
Family:PG1857-like |
|
|
| 88 | c1vjqB_ |
|
not modelled |
19.6 |
23 |
PDB header:structural genomics, de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
|
|
|
| 89 | c2bmaA_ |
|
not modelled |
18.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutamate dehydrogenase (nadp+);
PDBTitle: the crystal structure of plasmodium falciparum glutamate2 dehydrogenase, a putative target for novel antimalarial3 drugs
|
|
|
| 90 | c2vznA_ |
|
not modelled |
17.9 |
33 |
PDB header:allergen
Chain: A: PDB Molecule:venom allergen 3;
PDBTitle: crystal structure of the major allergen from fire ant venom, sol i 3
|
|
|
| 91 | c4ex8A_ |
|
not modelled |
17.8 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:alna;
PDBTitle: crystal structure of the prealnumycin c-glycosynthase alna
|
|
|
| 92 | c6bbmA_ |
|
not modelled |
17.4 |
15 |
PDB header:replication
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: mechanisms of opening and closing of the bacterial replicative2 helicase: the dnab helicase and lambda p helicase loader complex
|
|
|
| 93 | c3gndC_ |
|
not modelled |
16.9 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
|
|
| 94 | c5firH_ |
|
not modelled |
16.9 |
17 |
PDB header:hydrolase
Chain: H: PDB Molecule:paxt-1;
PDBTitle: crystal structure of c. elegans xrn2 in complex with the2 xrn2-binding domain of paxt-1
|
|
|
| 95 | d1qnxa_ |
|
not modelled |
16.9 |
30 |
Fold:PR-1-like
Superfamily:PR-1-like
Family:PR-1-like |
|
|
| 96 | d1pbya1 |
|
not modelled |
16.8 |
23 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
|
|
| 97 | c5weeD_ |
|
not modelled |
16.7 |
20 |
PDB header:lipid binding protein
Chain: D: PDB Molecule:venom allergen like protein 4;
PDBTitle: crystal structure of hpval4
|
|
|
| 98 | c3if8A_ |
|
not modelled |
16.4 |
13 |
PDB header:cell cycle
Chain: A: PDB Molecule:protein zwilch homolog;
PDBTitle: crystal structure of zwilch, a member of the rzz kinetochore complex
|
|
|
| 99 | c2vyeA_ |
|
not modelled |
15.8 |
15 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of the dnac-ssdna complex
|
|
|