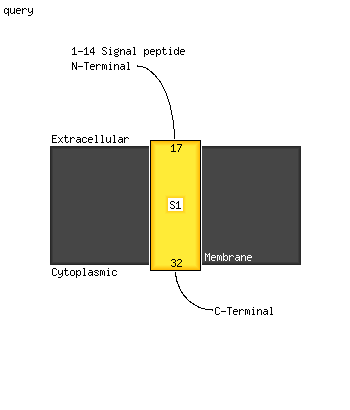
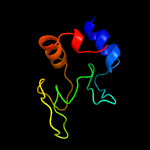
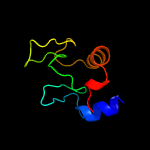
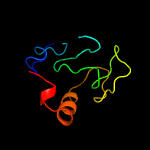
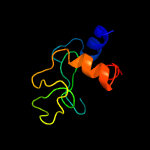
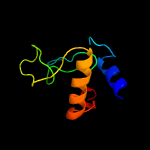
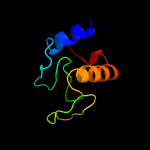
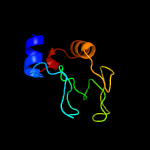
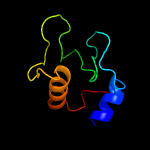
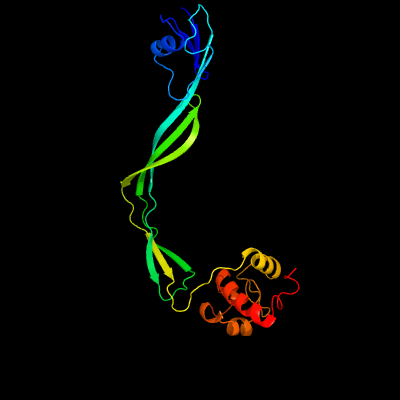1 c5e27B_
100.0
100
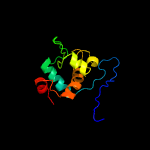
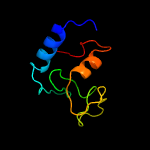
PDB header: cell adhesionChain: B: PDB Molecule: resuscitation-promoting factor rpfb;PDBTitle: the structure of resuscitation promoting factor b from m. tuberculosis2 reveals unexpected ubiquitin-like domains
2 c3eo5A_
100.0
96
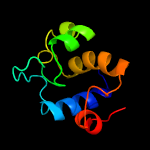
PDB header: cell adhesionChain: A: PDB Molecule: resuscitation-promoting factor rpfb;PDBTitle: crystal structure of the resuscitation promoting factor rpfb
3 c1xsfA_
100.0
100
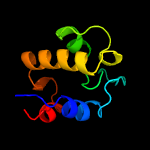
PDB header: cell cycle, hydrolaseChain: A: PDB Molecule: probable resuscitation-promoting factor rpfb;PDBTitle: solution structure of a resuscitation promoting factor2 domain from mycobacterium tuberculosis
4 d1xsfa1
100.0
100
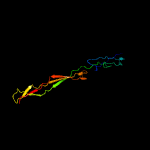
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: RPF-like
5 c4ow1A_
100.0
54
PDB header: hydrolaseChain: A: PDB Molecule: resuscitation-promoting factor rpfc;PDBTitle: crystal structure of resuscitation promoting factor c
6 c3tipA_
99.8
14
PDB header: structural proteinChain: A: PDB Molecule: surface protein g;PDBTitle: crystal structure of staphylococcus aureus sasg e-g52 module
7 c2ltjA_
99.8
20
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: conformational analysis of strh, the surface-attached exo- beta-d-n-2 acetylglucosaminidase from streptococcus pneumoniae
8 c4fzqC_
99.7
14
PDB header: immune systemChain: C: PDB Molecule: uncharacterized protein conserved in bacteria;PDBTitle: crystal structure of hp0197-g5
9 c4fuoA_
99.6
20
PDB header: membrane proteinChain: A: PDB Molecule: accumulation associated protein;PDBTitle: structural basis for zn2+-dependent intercellular adhesion in2 staphylococcal biofilms
10 c3tiqB_
99.5
13
PDB header: structural proteinChain: B: PDB Molecule: surface protein g;PDBTitle: crystal structure of staphylococcus aureus sasg g51-e-g52 module
11 d1hfxa_
97.6
22
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
12 c2fbdB_
97.6
24
PDB header: hydrolaseChain: B: PDB Molecule: lysozyme 1;PDBTitle: the crystallographic structure of the digestive lysozyme 1 from musca2 domestica at 1.90 ang.
13 d1yroa1
97.6
18
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
14 d1f6sa_
97.5
15
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
15 d1fkqa_
97.5
18
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
16 d1b9oa_
97.5
19
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
17 c2z2fA_
97.4
26
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme c-2;PDBTitle: x-ray crystal structure of bovine stomach lysozyme
18 d1alca_
97.3
17
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
19 d1iiza_
97.3
22
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
20 d1jsea_
97.0
18
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
21 d1hhla_
not modelled
97.0
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
22 d1gd6a_
not modelled
97.0
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
23 d2vb1a1
not modelled
96.9
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
24 d1ghla_
not modelled
96.9
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
25 d1ivma_
not modelled
96.8
27
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
26 d1lsga1
not modelled
96.8
18
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
27 d1qqya_
not modelled
96.8
22
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
28 d2eqla_
not modelled
96.7
23
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
29 d1juga_
not modelled
96.6
23
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
30 d1lmqa_
not modelled
96.5
23
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
31 d2nwdx1
not modelled
96.5
28
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: C-type lysozyme
32 c2goiC_
not modelled
96.5
21
PDB header: cell adhesion, sugar binding proteinChain: C: PDB Molecule: sperm lysozyme-like protein 1;PDBTitle: crystal structure of mouse sperm c-type lysozyme-like protein 1
33 d1qsaa2
not modelled
96.4
23
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Bacterial muramidase, catalytic domain
34 c3w6dB_
not modelled
96.4
13
PDB header: hydrolaseChain: B: PDB Molecule: lysozyme-like chitinolytic enzyme;PDBTitle: crystal structure of catalytic domain of chitinase from ralstonia sp.2 a-471 (e141q) in complex with tetrasaccharide
35 c2y8pA_
not modelled
96.0
22
PDB header: lyaseChain: A: PDB Molecule: endo-type membrane-bound lytic murein transglycosylase a;PDBTitle: crystal structure of an outer membrane-anchored endolytic2 peptidoglycan lytic transglycosylase (mlte) from3 escherichia coli
36 c3bkhA_
not modelled
95.9
27
PDB header: hydrolaseChain: A: PDB Molecule: lytic transglycosylase;PDBTitle: crystal structure of the bacteriophage phikz lytic2 transglycosylase, gp144
37 c4cfoB_
not modelled
95.9
19
PDB header: hydrolaseChain: B: PDB Molecule: mltc;PDBTitle: structure of lytic transglycosylase mltc from escherichia2 coli in complex with tetrasaccharide at 2.9 a resolution.
38 d1gbsa_
not modelled
94.1
23
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: G-type lysozyme
39 c4xp8A_
not modelled
93.9
20
PDB header: hydrolaseChain: A: PDB Molecule: etga protein;PDBTitle: structure of etga d60n mutant
40 c4oz9A_
not modelled
93.8
19
PDB header: lyaseChain: A: PDB Molecule: membrane-bound lytic murein transglycosylase f;PDBTitle: crystal structure of mltf from pseudomonas aeruginosa complexed with2 isoleucine
41 c3gxkB_
not modelled
93.4
22
PDB header: hydrolaseChain: B: PDB Molecule: goose-type lysozyme 1;PDBTitle: the crystal structure of g-type lysozyme from atlantic cod (gadus2 morhua l.) in complex with nag oligomers sheds new light on substrate3 binding and the catalytic mechanism. native structure to 1.9
42 c3mgwA_
not modelled
91.1
15
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme g;PDBTitle: thermodynamics and structure of a salmon cold-active goose-type2 lysozyme
43 c6cfcA_
not modelled
90.4
15
PDB header: hydrolaseChain: A: PDB Molecule: lytic transglycosylase;PDBTitle: crystal structure of soluble lytic transglycosylase cj0843 of2 campylobacter jejuni in complex with bulgecin a
44 d1y7ma2
not modelled
90.1
9
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain
45 c6fcqA_
not modelled
87.1
20
PDB header: lyaseChain: A: PDB Molecule: soluble lytic murein transglycosylase;PDBTitle: the x-ray structure of lytic transglycosylase slt inactive mutant2 e503q from pseudomonas aeruginosa in complex with bulgecin a
46 c4ebzA_
not modelled
85.6
15
PDB header: transferaseChain: A: PDB Molecule: chitin elicitor receptor kinase 1;PDBTitle: crystal structure of the ectodomain of a receptor like kinase
47 c1slyA_
not modelled
85.2
23
PDB header: glycosyltransferaseChain: A: PDB Molecule: 70-kda soluble lytic transglycosylase;PDBTitle: complex of the 70-kda soluble lytic transglycosylase with2 bulgecin a
48 c5ls2B_
not modelled
83.1
9
PDB header: plant proteinChain: B: PDB Molecule: lysm type receptor kinase;PDBTitle: receptor mediated chitin perception in legumes is functionally2 seperable from nod factor perception
49 c5k2lA_
not modelled
83.0
16
PDB header: hydrolaseChain: A: PDB Molecule: chitinase, lysozyme;PDBTitle: crystal structure of lysm domain from volvox carteri chitinase
50 c2mkxA_
not modelled
82.5
13
PDB header: hydrolaseChain: A: PDB Molecule: autolysin;PDBTitle: solution structure of lysm the peptidoglycan binding domain of2 autolysin atla from enterococcus faecalis
51 d1tygb_
not modelled
82.4
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS
52 c2kl0A_
not modelled
80.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
53 d2cu3a1
not modelled
79.1
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS
54 c3ct5A_
not modelled
78.2
16
PDB header: hydrolaseChain: A: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall degrading enzyme2 in the bacteriophage phi29 tail
55 c4pxvC_
not modelled
76.0
11
PDB header: sugar binding proteinChain: C: PDB Molecule: chitinase a;PDBTitle: crystal structure of lysm domain from pteris ryukyuensis chitinase a
56 c5bumA_
not modelled
75.5
12
PDB header: sugar binding proteinChain: A: PDB Molecule: chitinase a;PDBTitle: crystal structure of lysm domain from equisetum arvense chitinase a
57 c5jceA_
not modelled
75.3
16
PDB header: sugar binding proteinChain: A: PDB Molecule: chitin elicitor-binding protein;PDBTitle: crystal structure of oscebip complex
58 c3l53F_
not modelled
75.0
20
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative fumarylacetoacetate isomerase/hydrolase;PDBTitle: crystal structure of a putative fumarylacetoacetate2 isomerase/hydrolase from oleispira antarctica
59 c4uz2D_
not modelled
74.8
13
PDB header: hydrolaseChain: D: PDB Molecule: cell wall-binding endopeptidase-related protein;PDBTitle: crystal structure of the n-terminal lysm domains from the putative2 nlpc/p60 d,l endopeptidase from t. thermophilus
60 c2l9yA_
not modelled
74.1
13
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
61 c1tygG_
not modelled
73.9
14
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
62 c4b9hA_
not modelled
72.9
15
PDB header: sugar binding proteinChain: A: PDB Molecule: extracellular protein 6;PDBTitle: cladosporium fulvum lysm effector ecp6 in complex with a2 beta-1,4-linked n-acetyl-d-glucosamine tetramer: i3c heavy3 atom derivative
63 d1e0ga_
not modelled
72.4
11
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain
64 c4xcmB_
not modelled
65.0
7
PDB header: hydrolaseChain: B: PDB Molecule: cell wall-binding endopeptidase-related protein;PDBTitle: crystal structure of the putative nlpc/p60 d,l endopeptidase from t.2 thermophilus
65 d1qusa_
not modelled
64.6
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Bacterial muramidase, catalytic domain
66 d1zud21
not modelled
63.0
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS
67 c3cwiA_
not modelled
62.3
19
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
68 c1i7oC_
not modelled
62.2
17
PDB header: isomerase, lyaseChain: C: PDB Molecule: 4-hydroxyphenylacetate degradation bifunctionalPDBTitle: crystal structure of hpce
69 c2dfuB_
not modelled
56.6
16
PDB header: isomeraseChain: B: PDB Molecule: probable 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from thermus thermophilus hb8
70 c4anrA_
not modelled
53.2
21
PDB header: lyaseChain: A: PDB Molecule: soluble lytic transglycosylase b;PDBTitle: crystal structure of soluble lytic transglycosylase sltb12 from pseudomonas aeruginosa
71 c4s3kA_
not modelled
53.0
6
PDB header: hydrolaseChain: A: PDB Molecule: spore germination protein yaah;PDBTitle: crystal structure of the bacillus megaterium qm b1551 spore cortex-2 lytic enzyme slel
72 d1sawa_
not modelled
51.5
17
Fold: FAHSuperfamily: FAHFamily: FAH
73 c5lc5G_
not modelled
49.0
22
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-ubiquinone oxidoreductase 75 kda subunit,PDBTitle: structure of mammalian respiratory complex i, class2
74 c2djpA_
not modelled
47.2
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
75 d1eh9a2
not modelled
44.5
30
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
76 c5anzA_
not modelled
44.0
23
PDB header: hydrolaseChain: A: PDB Molecule: soluble lytic transglycosylase b3;PDBTitle: crystal structure of sltb3 from pseudomonas aeruginosa.
77 c6jvwA_
not modelled
43.9
13
PDB header: hydrolaseChain: A: PDB Molecule: maleylpyruvate hydrolase;PDBTitle: crystal structure of maleylpyruvate hydrolase from sphingobium sp.2 syk-6 in complex with manganese (ii) ion and pyruvate
78 d3c8ya2
not modelled
41.5
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
79 d1nr9a_
not modelled
41.5
16
Fold: FAHSuperfamily: FAHFamily: FAH
80 c1fuiB_
not modelled
40.9
33
PDB header: isomeraseChain: B: PDB Molecule: l-fucose isomerase;PDBTitle: l-fucose isomerase from escherichia coli
81 c4s3jC_
not modelled
40.4
16
PDB header: hydrolaseChain: C: PDB Molecule: cortical-lytic enzyme;PDBTitle: crystal structure of the bacillus cereus spore cortex-lytic enzyme2 slel
82 c4nooA_
not modelled
40.2
46
PDB header: immune systemChain: A: PDB Molecule: vgrg protein;PDBTitle: molecular mechanism for self-protection against type vi secretion2 system in vibrio cholerae
83 c5xf9F_
not modelled
39.2
17
PDB header: oxidoreductaseChain: F: PDB Molecule: nad-reducing hydrogenase;PDBTitle: crystal structure of nad+-reducing [nife]-hydrogenase in the air-2 oxidized state
84 c3u7zA_
not modelled
37.4
22
PDB header: metal binding proteinChain: A: PDB Molecule: putative metal binding protein rumgna_00854;PDBTitle: crystal structure of a putative metal binding protein rumgna_008542 (zp_02040092.1) from ruminococcus gnavus atcc 29149 at 1.30 a3 resolution
85 c1y7mB_
not modelled
34.5
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein bsu14040;PDBTitle: crystal structure of the b. subtilis ykud protein at 2 a2 resolution
86 c3csqC_
not modelled
33.2
17
PDB header: hydrolaseChain: C: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall2 degrading enzyme in the bacteriophage phi29 tail
87 c5fimA_
not modelled
32.8
5
PDB header: unknown functionChain: A: PDB Molecule: ygau;PDBTitle: the structure of kbp.k from e. coli
88 d1gtta1
not modelled
32.2
17
Fold: FAHSuperfamily: FAHFamily: FAH
89 c5ldxG_
not modelled
32.0
24
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-ubiquinone oxidoreductase 75 kda subunit,PDBTitle: structure of mammalian respiratory complex i, class3.
90 c5ldwG_
not modelled
32.0
24
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-ubiquinone oxidoreductase 75 kda subunit,PDBTitle: structure of mammalian respiratory complex i, class1
91 d1nkqa_
not modelled
30.0
19
Fold: FAHSuperfamily: FAHFamily: FAH
92 c5lnk3_
not modelled
29.8
22
PDB header: oxidoreductaseChain: 3: PDB Molecule: mitochondrial complex i, 75 kda subunit;PDBTitle: entire ovine respiratory complex i
93 c3qdfA_
not modelled
29.3
21
PDB header: isomeraseChain: A: PDB Molecule: 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from mycobacterium marinum
94 c1wzoC_
not modelled
29.2
19
PDB header: isomeraseChain: C: PDB Molecule: hpce;PDBTitle: crystal structure of the hpce from thermus thermophilus hb8
95 c4maqB_
not modelled
28.9
15
PDB header: hydrolaseChain: B: PDB Molecule: putative fumarylpyruvate hydrolase;PDBTitle: crystal structure of a putative fumarylpyruvate hydrolase from2 burkholderia cenocepacia
96 d2fug33
not modelled
27.3
28
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
97 d1gtta2
not modelled
26.6
16
Fold: FAHSuperfamily: FAHFamily: FAH
98 d1sv6a_
not modelled
25.8
11
Fold: FAHSuperfamily: FAHFamily: FAH
99 c3s52A_
not modelled
25.7
15
PDB header: hydrolaseChain: A: PDB Molecule: putative fumarylacetoacetate hydrolase family protein;PDBTitle: crystal structure of a putative fumarylacetoacetate hydrolase family2 protein from yersinia pestis co92
100 c3a9rA_
not modelled
22.7
32
PDB header: isomeraseChain: A: PDB Molecule: d-arabinose isomerase;PDBTitle: x-ray structures of bacillus pallidus d-arabinose2 isomerasecomplex with (4r)-2-methylpentane-2,4-diol
101 c6gcsA_
not modelled
22.2
22
PDB header: oxidoreductaseChain: A: PDB Molecule: 75-kda protein (nuam);PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
102 c6iymB_
not modelled
22.0
23
PDB header: hydrolaseChain: B: PDB Molecule: 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase;PDBTitle: fumarylacetoacetate hydrolase (eafah) from psychrophilic2 exiguobacterium antarcticum
103 c5j9hA_
not modelled
21.7
25
PDB header: viral proteinChain: A: PDB Molecule: envelopment polyprotein;PDBTitle: crystal structure of glycoprotein c from puumala virus in the post-2 fusion conformation (ph 8.0)