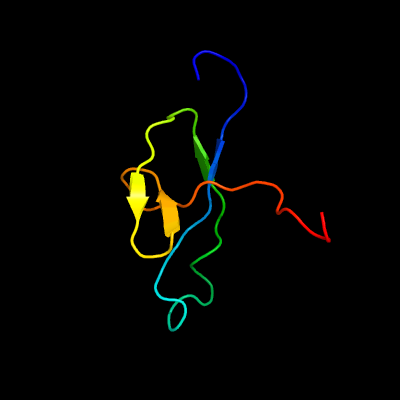
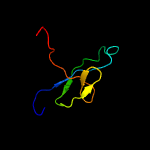
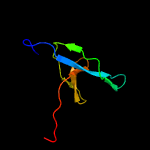
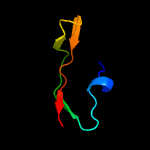
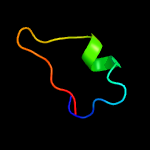
1 d1wlga_
18.5
16
Fold: Flagellar hook protein flgESuperfamily: Flagellar hook protein flgEFamily: Flagellar hook protein flgE
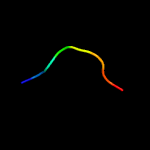
2 c5az4A_
17.8
18
PDB header: motor proteinChain: A: PDB Molecule: flagellar hook subunit protein;PDBTitle: crystal structure of a 79kda fragment of flge, the hook protein from2 campylobacter jejuni
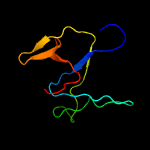
3 c4kh9B_
14.5
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a duf4785 family protein (lpg0956) from2 legionella pneumophila subsp. pneumophila str. philadelphia 1 at 2.003 a resolution
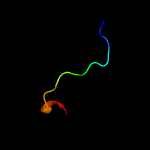
4 c3k0bA_
13.9
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted n6-adenine-specific dna methylase;PDBTitle: crystal structure of a predicted n6-adenine-specific dna methylase2 from listeria monocytogenes str. 4b f2365
5 d1ht6a1
12.8
24
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
6 d1o9ga_
12.7
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA methyltransferase AviRa
7 c3be3A_
11.7
63
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein belonging to pfam duf16532 from bordetella bronchiseptica
8 c5npyA_
11.2
8
PDB header: motor proteinChain: A: PDB Molecule: flagellar basal body protein;PDBTitle: crystal structure of helicobacter pylori flagellar hook protein flge2
9 c4js0B_
10.1
29
PDB header: signaling protein/signaling proteinChain: B: PDB Molecule: brain-specific angiogenesis inhibitor 1-associated proteinPDBTitle: complex of cdc42 with the crib-pr domain of irsp53
10 c3a0fA_
9.6
17
PDB header: hydrolaseChain: A: PDB Molecule: xyloglucanase;PDBTitle: the crystal structure of geotrichum sp. m128 xyloglucanase
11 c3v8vB_
9.2
43
PDB header: transferaseChain: B: PDB Molecule: ribosomal rna large subunit methyltransferase l;PDBTitle: crystal structure of bifunctional methyltransferase ycby (rlmlk) from2 escherichia coli, sam binding
12 c2mwrA_
9.1
50
PDB header: antimicrobial proteinChain: A: PDB Molecule: acidocin b;PDBTitle: solution structure of acidocin b, a circular bacteriocin from2 lactobacillus acidophilus m46
13 d1zdva1
9.1
60
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain
14 d1zdxa1
8.9
60
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain
15 c2cn2C_
8.4
10
PDB header: hydrolaseChain: C: PDB Molecule: beta-1,4-xyloglucan hydrolase;PDBTitle: crystal structures of clostridium thermocellum2 xyloglucanase
16 d2jdid2
8.4
41
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase
17 c2ebsB_
8.4
14
PDB header: hydrolaseChain: B: PDB Molecule: oligoxyloglucan reducing end-specificPDBTitle: crystal structure anaalysis of oligoxyloglucan reducing-end-2 specific cellobiohydrolase (oxg-rcbh) d465n mutant3 complexed with a xyloglucan heptasaccharide
18 c3nrqB_
8.3
31
PDB header: transport proteinChain: B: PDB Molecule: periplasmic protein-probably involved in high-affinity fe2+PDBTitle: crystal structure of copper-reconstituted fetp from uropathogenic2 escherichia coli strain f11
19 c5a6wC_
8.3
30
PDB header: antiviral proteinChain: C: PDB Molecule: avr-pik protein;PDBTitle: complex of rice blast (magnaporthe oryzae) effector protein avr-pikd2 with the hma domain of pikp1 from rice (oryza sativa)
20 c4parC_
7.9
8
PDB header: dna binding protein/dnaChain: C: PDB Molecule: uncharacterized protein abasi;PDBTitle: the 5-hydroxymethylcytosine-specific restriction enzyme abasi in a2 complex with product-like dna
21 d1pe4a_
not modelled
7.5
20
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Long-chain scorpion toxins
22 d1avaa1
not modelled
7.1
24
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
23 d2nn6h2
not modelled
6.9
22
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like
24 d1q42a_
not modelled
6.8
56
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: NTF2-like
25 c6ffyA_
not modelled
6.8
9
PDB header: apoptosisChain: A: PDB Molecule: vps10 domain-containing receptor sorcs2;PDBTitle: structure of the mouse sorcs2-ngf complex
26 c2xtlB_
not modelled
6.6
33
PDB header: structural proteinChain: B: PDB Molecule: cell wall surface anchor family protein;PDBTitle: structure of the major pilus backbone protein from streptococcus2 agalactiae
27 c3rpkA_
not modelled
6.5
40
PDB header: cell adhesionChain: A: PDB Molecule: backbone pilus subunit;PDBTitle: structure of the full-length major pilin rrgb from streptococcus2 pneumoniae
28 c6f0kD_
not modelled
6.3
15
PDB header: membrane proteinChain: D: PDB Molecule: actd;PDBTitle: alternative complex iii
29 d2ba0a2
not modelled
6.2
26
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like
30 d2adza1
not modelled
6.1
15
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
31 c2k9uB_
not modelled
6.0
75
PDB header: structural proteinChain: B: PDB Molecule: filamin-binding lim protein 1;PDBTitle: solution nmr structure of the filamin-migfilin complex
32 c6ftkA_
not modelled
5.9
9
PDB header: viral proteinChain: A: PDB Molecule: envelope protein;PDBTitle: gp36-mper
33 c3rlcA_
not modelled
5.9
25
PDB header: structural proteinChain: A: PDB Molecule: a1 protein;PDBTitle: crystal structure of the read-through domain from bacteriophage qbeta2 a1 protein, hexagonal crystal form
34 c1ikqA_
not modelled
5.8
18
PDB header: transferaseChain: A: PDB Molecule: exotoxin a;PDBTitle: pseudomonas aeruginosa exotoxin a, wild type
35 d1z0mb1
not modelled
5.8
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: AMPK-beta glycogen binding domain-like
36 c2qeyA_
not modelled
5.7
34
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxykinase, cytosolic [gtp];PDBTitle: rat cytosolic pepck in complex with gtp
37 c1q40C_
not modelled
5.7
56
PDB header: translationChain: C: PDB Molecule: mrna transport regulator mtr2;PDBTitle: crystal structure of the c. albicans mtr2-mex67 m domain complex
38 c3q8jA_
not modelled
5.4
33
PDB header: toxinChain: A: PDB Molecule: asteropsin a;PDBTitle: crystal structure of asteropsin a from marine sponge asteropus sp.
39 d3bwud1
not modelled
5.3
54
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain