| 1 | c5c4nD_
|
|
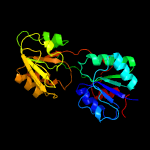 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:precorrin-6a reductase;
PDBTitle: cobk precorrin-6a reductase
|
|
|
|
| 2 | c4inaA_
|
|
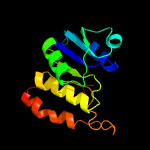 |
97.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: crystal structure of the q7mss8_wolsu protein from wolinella2 succinogenes. northeast structural genomics consortium target wsr35
|
|
|
|
| 3 | c4rl6A_
|
|
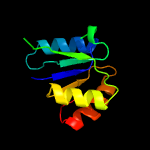 |
97.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: crystal structure of the q04l03_strp2 protein from streptococcus2 pneumoniae. northeast structural genomics consortium target spr105
|
|
|
|
| 4 | c2qx7A_
|
|
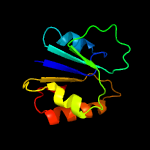 |
97.2 |
16 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
|
|
|
| 5 | c2dwcB_
|
|
 |
97.2 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
|
|
|
| 6 | c1gsoA_
|
|
 |
97.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:protein (glycinamide ribonucleotide synthetase);
PDBTitle: glycinamide ribonucleotide synthetase (gar-syn) from e.2 coli.
|
|
|
|
| 7 | c5z2fA_
|
|
 |
97.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
|
|
|
|
| 8 | c2qk4A_
|
|
 |
97.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:trifunctional purine biosynthetic protein adenosine-3;
PDBTitle: human glycinamide ribonucleotide synthetase
|
|
|
|
| 9 | c4ywjB_
|
|
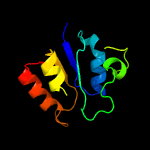 |
97.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate reductase (htpa2 reductase) from pseudomonas aeruginosa
|
|
|
|
| 10 | c2z2vA_
|
|
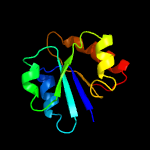 |
96.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
|
|
|
| 11 | c1kjjA_
|
|
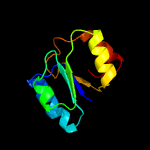 |
96.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide transformylase in2 complex with mg-atp-gamma-s
|
|
|
|
| 12 | c3lp8A_
|
|
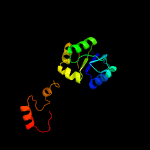 |
96.9 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine-glycine ligase;
PDBTitle: crystal structure of phosphoribosylamine-glycine ligase from2 ehrlichia chaffeensis
|
|
|
|
| 13 | c2ip4A_
|
|
 |
96.8 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of glycinamide ribonucleotide synthetase from2 thermus thermophilus hb8
|
|
|
|
| 14 | c2xd4A_
|
|
 |
96.8 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
|
|
|
|
| 15 | c2yyaB_
|
|
 |
96.7 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of gar synthetase from aquifex aeolicus
|
|
|
|
| 16 | d1gsoa2
|
|
 |
96.5 |
25 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
|
| 17 | c3e48B_
|
|
 |
96.4 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of a nucleoside-diphosphate-sugar epimerase2 (sav0421) from staphylococcus aureus, northeast structural genomics3 consortium target zr319
|
|
|
|
| 18 | c3ic5A_
|
|
 |
96.4 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
|
|
|
| 19 | c4dimA_
|
|
 |
96.3 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylglycinamide synthetase;
PDBTitle: crystal structure of phosphoribosylglycinamide synthetase from2 anaerococcus prevotii
|
|
|
|
| 20 | c5ugjC_
|
|
 |
96.3 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
|
|
|
|
| 21 | c5tenH_ |
|
not modelled |
96.3 |
18 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
|
|
|
| 22 | c5vevB_ |
|
not modelled |
96.3 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of phosphoribosylamine-glycine ligase from neisseria2 gonorrhoeae
|
|
|
| 23 | d1m1nb_ |
|
not modelled |
96.2 |
19 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 24 | c5kt0A_ |
|
not modelled |
96.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 25 | c2ys6A_ |
|
not modelled |
96.2 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylglycinamide synthetase;
PDBTitle: crystal structure of gar synthetase from geobacillus kaustophilus
|
|
|
| 26 | c1e5lA_ |
|
not modelled |
96.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
|
|
| 27 | c3c1oA_ |
|
not modelled |
96.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol synthase;
PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
|
|
|
| 28 | c3dhnA_ |
|
not modelled |
96.0 |
13 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn from2 bacteroides thetaiotaomicron. northeast structural genomics3 consortium target btr310.
|
|
|
| 29 | d1qh8b_ |
|
not modelled |
96.0 |
14 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 30 | c5ua0B_ |
|
not modelled |
96.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase 2,
PDBTitle: dimeric crystal structure of htpa reductase from arabidopsis thaliana
|
|
|
| 31 | c5kojD_ |
|
not modelled |
95.9 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitrogenase femo beta subunit protein nifk;
PDBTitle: nitrogenase mofep protein in the ids oxidized state
|
|
|
| 32 | c2axqA_ |
|
not modelled |
95.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
|
|
| 33 | d1e6ua_ |
|
not modelled |
95.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 34 | d1i24a_ |
|
not modelled |
95.7 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 35 | c5mlkA_ |
|
not modelled |
95.7 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: biotin dependent carboxylase acca3 dimer from mycobacterium2 tuberculosis (rv3285)
|
|
|
| 36 | c4ffnA_ |
|
not modelled |
95.7 |
14 |
PDB header:ligase/substrate
Chain: A: PDB Molecule:pylc;
PDBTitle: pylc in complex with d-ornithine and amppnp
|
|
|
| 37 | c3i5mA_ |
|
not modelled |
95.5 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative leucoanthocyanidin reductase 1;
PDBTitle: structure of the apo form of leucoanthocyanidin reductase from vitis2 vinifera
|
|
|
| 38 | c2dzdB_ |
|
not modelled |
95.5 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of the biotin carboxylase domain of pyruvate2 carboxylase
|
|
|
| 39 | c2q1wC_ |
|
not modelled |
95.4 |
18 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
|
|
| 40 | c5ffqA_ |
|
not modelled |
95.4 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:shuy-like protein;
PDBTitle: chuy: an anaerobillin reductase from escherichia coli o157:h7
|
|
|
| 41 | c5l78A_ |
|
not modelled |
95.4 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-aminoadipic semialdehyde synthase, mitochondrial;
PDBTitle: crystal structure of human aminoadipate semialdehyde synthase,2 saccharopine dehydrogenase domain (in nad+ bound form)
|
|
|
| 42 | c3ouzA_ |
|
not modelled |
95.4 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
|
|
|
| 43 | d1qyca_ |
|
not modelled |
95.3 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 44 | c5l3zA_ |
|
not modelled |
95.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyketide ketoreductase simc7;
PDBTitle: polyketide ketoreductase simc7 - binary complex with nadp+
|
|
|
| 45 | c2gpwC_ |
|
not modelled |
95.2 |
7 |
PDB header:ligase
Chain: C: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of the biotin carboxylase subunit, f363a2 mutant, of acetyl-coa carboxylase from escherichia coli.
|
|
|
| 46 | d1m1na_ |
|
not modelled |
95.2 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 47 | c3nklA_ |
|
not modelled |
95.2 |
16 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
|
|
| 48 | c1yl7F_ |
|
not modelled |
95.2 |
20 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: the crystal structure of mycobacterium tuberculosis2 dihydrodipicolinate reductase (rv2773c) in complex with nadh (crystal3 form c)
|
|
|
| 49 | c1drwA_ |
|
not modelled |
95.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: escherichia coli dhpr/nhdh complex
|
|
|
| 50 | c4f3yA_ |
|
not modelled |
95.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: x-ray crystal structure of dihydrodipicolinate reductase from2 burkholderia thailandensis
|
|
|
| 51 | c3sc6F_ |
|
not modelled |
94.9 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
|
|
| 52 | c2yy7B_ |
|
not modelled |
94.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-threonine dehydrogenase;
PDBTitle: crystal structure of thermolabile l-threonine dehydrogenase from2 flavobacterium frigidimaris kuc-1
|
|
|
| 53 | c4wpgA_ |
|
not modelled |
94.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: group a streptococcus gaca is an essential dtdp-4-dehydrorhamnose2 reductase (rmld)
|
|
|
| 54 | c3g8cB_ |
|
not modelled |
94.8 |
8 |
PDB header:ligase
Chain: B: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase in complex with biotin,2 bicarbonate, adp and mg ion
|
|
|
| 55 | d1qh8a_ |
|
not modelled |
94.8 |
18 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 56 | c2ph5A_ |
|
not modelled |
94.7 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
|
|
| 57 | d1kjqa2 |
|
not modelled |
94.6 |
22 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 58 | d1xgka_ |
|
not modelled |
94.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 59 | c4plpB_ |
|
not modelled |
94.5 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase (hss) from2 blastochloris viridis in complex with nad
|
|
|
| 60 | d1bxka_ |
|
not modelled |
94.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 61 | c1ulzA_ |
|
not modelled |
94.4 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase n-terminal domain;
PDBTitle: crystal structure of the biotin carboxylase subunit of pyruvate2 carboxylase
|
|
|
| 62 | c1z7eC_ |
|
not modelled |
94.4 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
|
|
| 63 | c4qqrB_ |
|
not modelled |
94.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3,5-epimerase/4-reductase;
PDBTitle: structural insight into nucleotide rhamnose synthase/epimerase-2 reductase from arabidopsis thaliana
|
|
|
| 64 | d1hdoa_ |
|
not modelled |
94.3 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 65 | c1tcvB_ |
|
not modelled |
94.2 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:purine-nucleoside phosphorylase;
PDBTitle: crystal structure of the purine nucleoside phosphorylase2 from schistosoma mansoni in complex with non-detergent3 sulfobetaine 195 and acetate
|
|
|
| 66 | c4jgbB_ |
|
not modelled |
94.2 |
16 |
PDB header:protein binding
Chain: B: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of putative exported protein from burkholderia2 pseudomallei
|
|
|
| 67 | c3ijpA_ |
|
not modelled |
94.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
|
|
|
| 68 | c5l9aB_ |
|
not modelled |
94.2 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-threonine 3-dehydrogenase;
PDBTitle: l-threonine dehydrogenase from trypanosoma brucei.
|
|
|
| 69 | c4b8wB_ |
|
not modelled |
94.1 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-l-fucose synthase;
PDBTitle: crystal structure of human gdp-l-fucose synthase with bound nadp and2 gdp, tetragonal crystal form
|
|
|
| 70 | c3ax6C_ |
|
not modelled |
94.0 |
13 |
PDB header:ligase
Chain: C: PDB Molecule:phosphoribosylaminoimidazole carboxylase, atpase subunit;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide2 synthetase from thermotoga maritima
|
|
|
| 71 | c3wmxC_ |
|
not modelled |
94.0 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad dependent epimerase/dehydratase;
PDBTitle: gale-like l-threonine dehydrogenase from cupriavidus necator (holo2 form)
|
|
|
| 72 | c5wolA_ |
|
not modelled |
94.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase dapb from coxiella2 burnetii
|
|
|
| 73 | d1pjqa1 |
|
not modelled |
94.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 74 | c5cxsA_ |
|
not modelled |
94.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of isoform 2 of purine nucleoside phosphorylase2 complexed with mes
|
|
|
| 75 | c2ynmD_ |
|
not modelled |
93.9 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
|
|
|
| 76 | c3u9sA_ |
|
not modelled |
93.8 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:methylcrotonyl-coa carboxylase, alpha-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
|
|
|
| 77 | d2c5aa1 |
|
not modelled |
93.8 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 78 | d1yl7a1 |
|
not modelled |
93.8 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 79 | c6aqyD_ |
|
not modelled |
93.6 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gdp-l-fucose synthetase;
PDBTitle: crystal structure of a gdp-l-fucose synthetase from naegleria fowleri
|
|
|
| 80 | d1mioa_ |
|
not modelled |
93.5 |
18 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 81 | c2zklA_ |
|
not modelled |
93.5 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:capsular polysaccharide synthesis enzyme cap5f;
PDBTitle: crystal structure of capsular polysaccharide assembling protein capf2 from staphylococcus aureus
|
|
|
| 82 | c3jzfA_ |
|
not modelled |
93.5 |
7 |
PDB header:ligase
Chain: A: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase from e. coli in2 complex with benzimidazoles series
|
|
|
| 83 | c3gidB_ |
|
not modelled |
93.5 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: the biotin carboxylase (bc) domain of human acetyl-coa carboxylase 22 (acc2) in complex with soraphen a
|
|
|
| 84 | c5us6L_ |
|
not modelled |
93.5 |
21 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: structure of dihydrodipicolinate reductase from vibrio vulnificus2 bound to nadh and 2,6 pyridine dicarboxylic acid with intact3 polyhistidine tag
|
|
|
| 85 | c2p5uC_ |
|
not modelled |
93.5 |
22 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-epimerase2 complex with nad
|
|
|
| 86 | c2z1mC_ |
|
not modelled |
93.4 |
19 |
PDB header:lyase
Chain: C: PDB Molecule:gdp-d-mannose dehydratase;
PDBTitle: crystal structure of gdp-d-mannose dehydratase from aquifex aeolicus2 vf5
|
|
|
| 87 | c1yr3A_ |
|
not modelled |
93.4 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:xanthosine phosphorylase;
PDBTitle: escherichia coli purine nucleoside phosphorylase ii, the product of2 the xapa gene
|
|
|
| 88 | c3pdiB_ |
|
not modelled |
93.3 |
21 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|
|
|
| 89 | c5ifkC_ |
|
not modelled |
93.2 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: purine nucleoside phosphorylase
|
|
|
| 90 | c4glfA_ |
|
not modelled |
93.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:rsfp;
PDBTitle: crystal structure of methylthioadenosine phosphorylase sourced from an2 antarctic soil metagenomic library
|
|
|
| 91 | c2pk3B_ |
|
not modelled |
93.1 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
|
|
| 92 | c3rgqA_ |
|
not modelled |
93.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine phosphatase mitochondrial 1;
PDBTitle: crystal structure of ptpmt1 in complex with pi(5)p
|
|
|
| 93 | c5eesA_ |
|
not modelled |
92.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: crystal strcuture of dapb in complex with nadp+ from corynebacterium2 glutamicum
|
|
|
| 94 | d1n7ha_ |
|
not modelled |
92.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 95 | c4twrA_ |
|
not modelled |
92.7 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:nad binding site:nad-dependent epimerase/dehydratase:udp-
PDBTitle: structure of udp-glucose 4-epimerase from brucella abortus
|
|
|
| 96 | c6f2cK_ |
|
not modelled |
92.7 |
12 |
PDB header:lyase
Chain: K: PDB Molecule:methylglyoxal synthase;
PDBTitle: methylglyoxal synthase mgsa from bacillus subtilis
|
|
|
| 97 | d2q46a1 |
|
not modelled |
92.7 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 98 | c1z45A_ |
|
not modelled |
92.6 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:gal10 bifunctional protein;
PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces cerevisiae3 complexed with nad, udp-glucose, and galactose
|
|
|
| 99 | d1kyqa1 |
|
not modelled |
92.6 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 100 | c4egqD_ |
|
not modelled |
92.5 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of d-alanine-d-alanine ligase b from burkholderia2 pseudomallei
|
|
|
| 101 | d1db3a_ |
|
not modelled |
92.5 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 102 | d1wo8a1 |
|
not modelled |
92.2 |
16 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
|
|
| 103 | c3etjB_ |
|
not modelled |
92.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase atpase
PDBTitle: crystal structure e. coli purk in complex with mg, adp, and2 pi
|
|
|
| 104 | c4egjD_ |
|
not modelled |
91.9 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of d-alanine-d-alanine ligase from burkholderia2 xenovorans
|
|
|
| 105 | d1vl0a_ |
|
not modelled |
91.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 106 | d1kewa_ |
|
not modelled |
91.8 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 107 | c3ew7A_ |
|
not modelled |
91.7 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo0794 protein;
PDBTitle: crystal structure of the lmo0794 protein from listeria2 monocytogenes. northeast structural genomics consortium3 target lmr162.
|
|
|
| 108 | d1oc2a_ |
|
not modelled |
91.7 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 109 | d1diha1 |
|
not modelled |
91.7 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 110 | c2p4sA_ |
|
not modelled |
91.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
|
|
|
| 111 | c4zrmB_ |
|
not modelled |
91.4 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of udp-glucose 4-epimerase (tm0509) from2 hyperthermophilic eubacterium thermotoga maritima
|
|
|
| 112 | c2yvqA_ |
|
not modelled |
91.4 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
|
|
| 113 | c3tw6B_ |
|
not modelled |
91.1 |
9 |
PDB header:ligase/activator
Chain: B: PDB Molecule:pyruvate carboxylase protein;
PDBTitle: structure of rhizobium etli pyruvate carboxylase t882a with the2 allosteric activator, acetyl coenzyme-a
|
|
|
| 114 | c3hblA_ |
|
not modelled |
91.1 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of s. aureus pyruvate carboxylase t908a mutant
|
|
|
| 115 | c1t2aC_ |
|
not modelled |
90.9 |
17 |
PDB header:structural genomics,lyase
Chain: C: PDB Molecule:gdp-mannose 4,6 dehydratase;
PDBTitle: crystal structure of human gdp-d-mannose 4,6-dehydratase
|
|
|
| 116 | d1t2aa_ |
|
not modelled |
90.9 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 117 | c3k5iB_ |
|
not modelled |
90.9 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosyl-aminoimidazole carboxylase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole synthase from2 aspergillus clavatus in complex with adp and 5-aminoimadazole3 ribonucleotide
|
|
|
| 118 | d2fmua1 |
|
not modelled |
90.9 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 119 | c4yraD_ |
|
not modelled |
90.8 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:l-threonine 3-dehydrogenase, mitochondrial;
PDBTitle: mouse tdh in the apo form
|
|
|
| 120 | c2xdqB_ |
|
not modelled |
90.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
|
|
|








































































































































































