| 1 | c6h59B_
|
|
 |
100.0 |
100 |
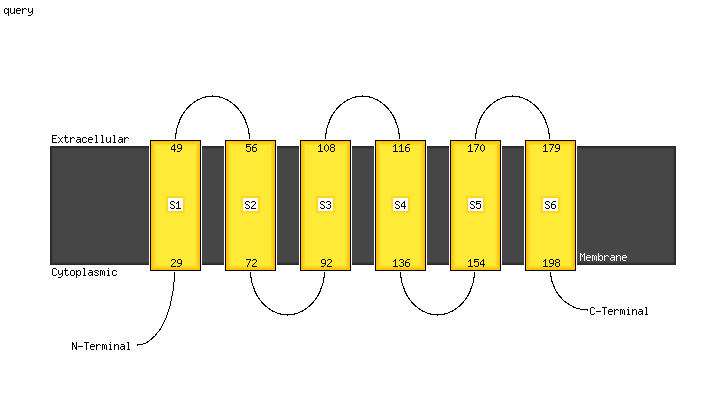
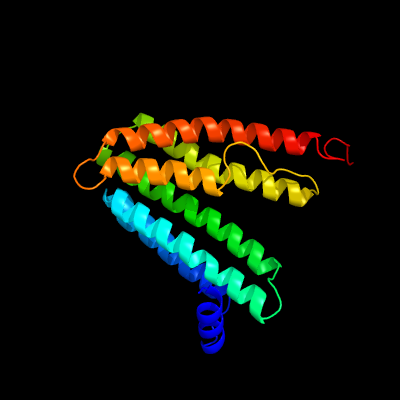
PDB header:transferase
Chain: B: PDB Molecule:cdp-diacylglycerol--inositol 3-phosphatidyltransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphatidylinositol2 phosphate synthase (pgsa1) with cdp-dag bound
|
|
|
|
| 2 | c5d92B_
|
|
 |
100.0 |
43 |
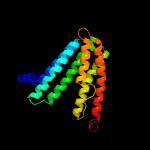
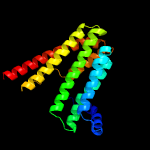
PDB header:membrane protein
Chain: B: PDB Molecule:af2299 protein,phosphatidylinositol synthase;
PDBTitle: structure of a phosphatidylinositolphosphate (pip) synthase from2 renibacterium salmoninarum
|
|
|
|
| 3 | c4o6mA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:af2299, a cdp-alcohol phosphotransferase;
PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
|
|
|
|
| 4 | c4mndA_
|
|
 |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:ctp l-myo-inositol-1-phosphate cytidylyltransferase/cdp-l-
PDBTitle: crystal structure of archaeoglobus fulgidus ipct-dipps bifunctional2 membrane protein
|
|
|
|
| 5 | c2voyG_
|
|
 |
52.3 |
17 |
PDB header:hydrolase
Chain: G: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium atpase 1;
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
|
|
|
|
| 6 | c5gufA_
|
|
 |
26.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:cdp-archaeol synthase;
PDBTitle: structural insight into an intramembrane enzyme for archaeal membrane2 lipids biosynthesis
|
|
|
|
| 7 | c5zc2B_
|
|
 |
25.4 |
13 |
PDB header:flavoprotein
Chain: B: PDB Molecule:p-hydroxyphenylacetate 3-hydroxylase, reductase component;
PDBTitle: acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (hpah),2 reductase component (c1)
|
|
|
|
| 8 | d1ou8a_
|
|
 |
22.6 |
31 |
Fold:SspB-like
Superfamily:SspB-like
Family:Stringent starvation protein B, SspB |
|
|
|
| 9 | d1zszc1
|
|
 |
22.2 |
19 |
Fold:SspB-like
Superfamily:SspB-like
Family:Stringent starvation protein B, SspB |
|
|
|
| 10 | d1ou9a_
|
|
 |
20.1 |
31 |
Fold:SspB-like
Superfamily:SspB-like
Family:Stringent starvation protein B, SspB |
|
|
|
| 11 | c2rkhA_
|
|
 |
19.1 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:putative apha-like transcription factor;
PDBTitle: crystal structure of a putative apha-like transcription factor2 (zp_00208345.1) from magnetospirillum magnetotacticum ms-1 at 2.00 a3 resolution
|
|
|
|
| 12 | d1yfna1
|
|
 |
18.2 |
25 |
Fold:SspB-like
Superfamily:SspB-like
Family:Stringent starvation protein B, SspB |
|
|
|
| 13 | c6mjbC_
|
|
 |
16.0 |
28 |
PDB header:cell cycle
Chain: C: PDB Molecule:kinetochore-associated protein dsn1;
PDBTitle: structure of candida glabrata csm1:dsn1(14-72) complex
|
|
|
|
| 14 | c3cjnA_
|
|
 |
13.0 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of transcriptional regulator, marr family, from2 silicibacter pomeroyi
|
|
|
|
| 15 | c6c28C_
|
|
 |
12.0 |
21 |
PDB header:dna binding protein
Chain: C: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: transcriptional repressor, cour, bound to p-coumaroyl-coa
|
|
|
|
| 16 | c3cdhB_
|
|
 |
11.5 |
16 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of the marr family transcriptional regulator spo14532 from silicibacter pomeroyi dss-3
|
|
|
|
| 17 | d2fbha1
|
|
 |
10.3 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
|
|
|
| 18 | c4n3xD_
|
|
 |
9.8 |
64 |
PDB header:endocytosis
Chain: D: PDB Molecule:rab5 gdp/gtp exchange factor;
PDBTitle: crystal structure of rabex-5 cc domain
|
|
|
|
| 19 | c2zxqA_
|
|
 |
9.6 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-alpha-n-acetylgalactosaminidase;
PDBTitle: crystal structure of endo-alpha-n-acetylgalactosaminidase2 from bifidobacterium longum (engbf)
|
|
|
|
| 20 | c3ecqA_
|
|
 |
9.2 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-alpha-n-acetylgalactosaminidase;
PDBTitle: endo-alpha-n-acetylgalactosaminidase from streptococcus pneumoniae:2 semet structure
|
|
|
|
| 21 | c4i0xJ_ |
|
not modelled |
8.5 |
22 |
PDB header:structural genomics, unknown function
Chain: J: PDB Molecule:esat-6-like protein mab_3113;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
|
|
|
| 22 | c3bj6B_ |
|
not modelled |
6.8 |
29 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcription regulator sp03579
|
|
|
| 23 | d1bvp11 |
|
not modelled |
6.7 |
29 |
Fold:A virus capsid protein alpha-helical domain
Superfamily:A virus capsid protein alpha-helical domain
Family:Orbivirus capsid |
|
|
| 24 | c3hrmA_ |
|
not modelled |
5.9 |
22 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator sarz;
PDBTitle: crystal structure of staphylococcus aureus protein sarz in sulfenic2 acid form
|
|
|
| 25 | c2le7A_ |
|
not modelled |
5.6 |
29 |
PDB header:transport protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h member 2;
PDBTitle: solution nmr structure of the s4s5 linker of herg potassium channel
|
|
|
| 26 | c3ecoB_ |
|
not modelled |
5.4 |
16 |
PDB header:transcription
Chain: B: PDB Molecule:mepr;
PDBTitle: crystal structure of mepr, a transcription regulator of the2 staphylococcus aureus multidrug efflux pump mepa
|
|
|
| 27 | c3oopA_ |
|
not modelled |
5.4 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2960 protein;
PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
|
|
|