| 1 |
|
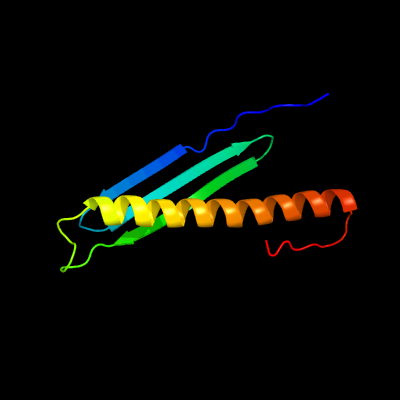
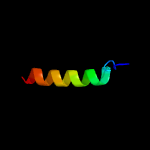
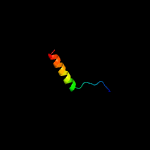
PDB 4wpy chain A
Region: 5 - 92
Aligned: 88
Modelled: 88
Confidence: 100.0%
Identity: 53%
PDB header:de novo protein
Chain: A: PDB Molecule:protein dl-rv1738;
PDBTitle: racemic crystal structure of rv1738 from mycobacterium tuberculosis2 (form-ii)
Phyre2
| 2 |
|
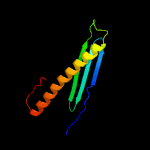
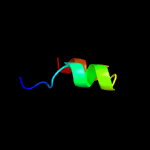
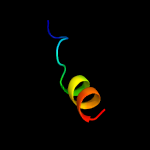
PDB 2fgg chain A domain 1
Region: 4 - 87
Aligned: 84
Modelled: 84
Confidence: 100.0%
Identity: 99%
Fold: dsRBD-like
Superfamily: Rv2632c-like
Family: Rv2632c-like
Phyre2
| 3 |
|
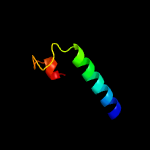
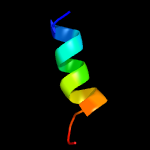
PDB 6j9m chain A
Region: 51 - 78
Aligned: 28
Modelled: 28
Confidence: 53.2%
Identity: 32%
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:crispr-associated endonuclease cas9;
PDBTitle: nmebh+acriic2
Phyre2
| 4 |
|
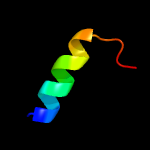
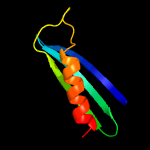
PDB 6j9m chain F
Region: 51 - 74
Aligned: 24
Modelled: 24
Confidence: 51.6%
Identity: 38%
PDB header:hydrolase/hydrolase inhibitor
Chain: F: PDB Molecule:crispr-associated endonuclease cas9;
PDBTitle: nmebh+acriic2
Phyre2
| 5 |
|
PDB 5mhj chain A
Region: 50 - 70
Aligned: 21
Modelled: 21
Confidence: 17.5%
Identity: 29%
PDB header:transcription
Chain: A: PDB Molecule:major viral transcription factor icp4;
PDBTitle: icp4 dna-binding domain, lacking intrinsically disordered region, in2 complex with 12mer dna duplex from its own promoter
Phyre2
| 6 |
|
PDB 2hjd chain A
Region: 57 - 92
Aligned: 36
Modelled: 36
Confidence: 15.6%
Identity: 25%
PDB header:signaling protein
Chain: A: PDB Molecule:quorum-sensing antiactivator;
PDBTitle: crystal structure of a second quorum sensing antiactivator tram2 from2 a. tumefaciens strain a6
Phyre2
| 7 |
|
PDB 1grj chain A domain 1
Region: 53 - 69
Aligned: 17
Modelled: 17
Confidence: 13.9%
Identity: 35%
Fold: Long alpha-hairpin
Superfamily: GreA transcript cleavage protein, N-terminal domain
Family: GreA transcript cleavage protein, N-terminal domain
Phyre2
| 8 |
|
PDB 6jdx chain C
Region: 48 - 74
Aligned: 27
Modelled: 27
Confidence: 11.9%
Identity: 33%
PDB header:hydrolase/hydrolase inhibitor
Chain: C: PDB Molecule:crispr-associated endonuclease cas9;
PDBTitle: crystal structure of acriic2 dimer in complex with partial nme1cas92 preprocessed with protease alpha-chymotrypsin
Phyre2
| 9 |
|
PDB 2f23 chain A domain 1
Region: 53 - 69
Aligned: 17
Modelled: 17
Confidence: 9.9%
Identity: 18%
Fold: Long alpha-hairpin
Superfamily: GreA transcript cleavage protein, N-terminal domain
Family: GreA transcript cleavage protein, N-terminal domain
Phyre2
| 10 |
|
PDB 4kzs chain A
Region: 58 - 72
Aligned: 15
Modelled: 15
Confidence: 5.6%
Identity: 33%
PDB header:unknown function
Chain: A: PDB Molecule:lpp20 lipofamily protein;
PDBTitle: crystal structure of the secreted protein hp1454 from the human2 pathogen helicobacter pylori
Phyre2
| 11 |
|
PDB 2voi chain B
Region: 52 - 59
Aligned: 8
Modelled: 8
Confidence: 5.6%
Identity: 75%
PDB header:apoptosis
Chain: B: PDB Molecule:bh3-interacting domain death agonist p13;
PDBTitle: structure of mouse a1 bound to the bid bh3-domain
Phyre2
| 12 |
|
PDB 2rcd chain A domain 1
Region: 47 - 62
Aligned: 16
Modelled: 16
Confidence: 5.2%
Identity: 25%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: BxeB1374-like
Phyre2
| 13 |
|
PDB 3ctd chain A domain 1
Region: 68 - 83
Aligned: 16
Modelled: 16
Confidence: 5.2%
Identity: 19%
Fold: post-AAA+ oligomerization domain-like
Superfamily: post-AAA+ oligomerization domain-like
Family: MgsA/YrvN C-terminal domain-like
Phyre2
| 14 |
|
PDB 1ci4 chain A
Region: 50 - 60
Aligned: 11
Modelled: 11
Confidence: 5.2%
Identity: 36%
Fold: SAM domain-like
Superfamily: Barrier-to-autointegration factor, BAF
Family: Barrier-to-autointegration factor, BAF
Phyre2
| 15 |
|
PDB 4fyl chain B
Region: 11 - 74
Aligned: 53
Modelled: 59
Confidence: 5.1%
Identity: 25%
PDB header:translation
Chain: B: PDB Molecule:ribosome hibernation protein yhbh;
PDBTitle: high-resolution x-ray structure of hpf from vibrio cholerae
Phyre2