1 c6ig0B_
100.0
38
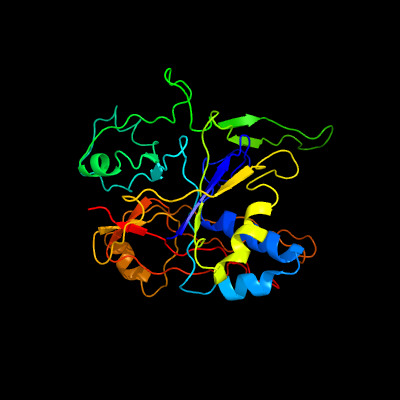
PDB header: rna binding proteinChain: B: PDB Molecule: type iii-a crispr-associated ramp protein csm4;PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
2 c6muuE_
100.0
23
PDB header: rna binding protein/rnaChain: E: PDB Molecule: uncharacterized protein csm4;PDBTitle: cryo-em structure of csm-crrna binary complex in type iii-a crispr-cas2 system
3 c4qtsB_
100.0
21
PDB header: rna binding proteinChain: B: PDB Molecule: crispr type iii-associated ramp protein csm4;PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
4 c4n0lB_
98.6
14
PDB header: rna binding proteinChain: B: PDB Molecule: predicted component of a thermophile-specific dna repairPDBTitle: methanopyrus kandleri csm3 crystal structure
5 c6ae2B_
98.6
22
PDB header: rna binding proteinChain: B: PDB Molecule: csm3;PDBTitle: crystal structure of csm3 of the type iii-a crispr-cas effector2 complex
6 c6ifnF_
98.5
16
PDB header: rna binding proteinChain: F: PDB Molecule: type iii-a crispr-associated ramp protein csm3;PDBTitle: crystal structure of type iii-a crispr csm complex
7 c4w8wB_
98.5
18
PDB header: hydrolaseChain: B: PDB Molecule: crispr system cmr subunit cmr4;PDBTitle: crystal structure of oligomeric cmr4 from pyrococcus furiosus
8 c5yjdB_
98.2
15
PDB header: hydrolaseChain: B: PDB Molecule: csm3;PDBTitle: structural insights into the crispr-cas-associated ribonuclease2 activity of staphylococcus epidermidis csm3
9 c6musK_
98.1
15
PDB header: rna binding protein/rnaChain: K: PDB Molecule: uncharacterized protein csm3;PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
10 c4w8vA_
97.4
13
PDB header: rna binding proteinChain: A: PDB Molecule: crispr system cmr subunit cmr6;PDBTitle: crystal structure of cmr6 from pyrococcus furiosus
11 c3x1lE_
97.3
19
PDB header: rna binding protein/rna/dnaChain: E: PDB Molecule: cmr4;PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
12 c4h4kA_
97.3
14
PDB header: rna binding proteinChain: A: PDB Molecule: crispr system cmr subunit cmr3;PDBTitle: structure of the cmr2-cmr3 subcomplex of the cmr rna-silencing complex
13 c4qtsC_
97.1
16
PDB header: rna binding proteinChain: C: PDB Molecule: crispr type iii-associated ramp protein csm3;PDBTitle: crystal structure of csm3-csm4 subcomplex in the type iii-a crispr-cas2 interference complex
14 c3x1lH_
96.1
16
PDB header: rna binding protein/rna/dnaChain: H: PDB Molecule: cmr6;PDBTitle: crystal structure of the crispr-cas rna silencing cmr complex bound to2 a target analog
15 c4ilmH_
92.8
19
PDB header: hydrolase/rnaChain: H: PDB Molecule: crispr-associated endoribonuclease cas6 2;PDBTitle: crispr rna processing endoribonuclease
16 c6fjwA_
84.2
28
PDB header: hydrolaseChain: A: PDB Molecule: cas6 protein;PDBTitle: streptococcus thermophilus cas6
17 c4c97B_
67.7
18
PDB header: hydrolaseChain: B: PDB Molecule: cas6a;PDBTitle: cas6 (ttha0078) h37a mutant
18 c4c9dA_
62.5
21
PDB header: hydrolase/rnaChain: A: PDB Molecule: cas6b;PDBTitle: cas6 (tthb231) product complex
19 c3i4hX_
59.7
17
PDB header: hydrolaseChain: X: PDB Molecule: endoribonuclease;PDBTitle: crystal structure of cas6 in pyrococcus furiosus
20 d1q0qa3
51.8
19
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
21 c5kqoA_
not modelled
48.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from vibrio vulnificus
22 c6dd5B_
not modelled
47.9
23
PDB header: hydrolaseChain: B: PDB Molecule: mmb-1 cas6 fused to maltose binding protein,crispr-PDBTitle: crystal structure of the cas6 domain of marinomonas mediterranea mmb-12 cas6-rt-cas1 fusion protein
23 c2h5xA_
not modelled
47.2
15
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction atp-dependent dna helicase ruva;PDBTitle: ruva from mycobacterium tuberculosis
24 c5yi6A_
not modelled
46.7
19
PDB header: hydrolaseChain: A: PDB Molecule: crispr-associated endoribonuclease cas6 1;PDBTitle: crispr associated protein cas6
25 c2qtpA_
not modelled
42.2
36
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf1185 family protein (spo0826) from2 silicibacter pomeroyi dss-3 at 2.10 a resolution
26 d2qtpa1
not modelled
42.2
36
Fold: Bacillus chorismate mutase-likeSuperfamily: BB2672-likeFamily: BB2672-like
27 d3byqa1
not modelled
40.9
19
Fold: Bacillus chorismate mutase-likeSuperfamily: BB2672-likeFamily: BB2672-like
28 c3au9A_
not modelled
40.8
17
PDB header: isomerase/isomerase inhibitorChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of the quaternary complex-1 of an isomerase
29 c6ig0H_
not modelled
40.6
11
PDB header: rna binding proteinChain: H: PDB Molecule: type iii-a crispr-associated ramp protein csm5;PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
30 c2eghA_
not modelled
39.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
31 c3qjjB_
not modelled
37.9
19
PDB header: immune system/rnaChain: B: PDB Molecule: putative uncharacterized protein ph0350;PDBTitle: one ramp protein binding different rna substrates
32 d2b9da1
not modelled
30.3
18
Fold: E7 C-terminal domain-likeSuperfamily: E7 C-terminal domain-likeFamily: E7 C-terminal domain-like
33 c6musF_
not modelled
29.2
15
PDB header: rna binding protein/rnaChain: F: PDB Molecule: uncharacterized protein csm5;PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
34 c6njyB_
not modelled
25.7
17
PDB header: rna binding proteinChain: B: PDB Molecule: type iv crispr associated cas6 rna endonuclease;PDBTitle: type iv crispr associated rna endonuclease cas6 - apo form
35 c2jcyA_
not modelled
24.3
23
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium tuberculosis
36 c1d8lA_
not modelled
23.4
15
PDB header: gene regulationChain: A: PDB Molecule: protein (holliday junction dna helicase ruva);PDBTitle: e. coli holliday junction binding protein ruva nh2 region lacking2 domain iii
37 c5yzcB_
not modelled
23.3
22
PDB header: viral proteinChain: B: PDB Molecule: glycoprotein f1,measles virus fusion protein;PDBTitle: crystal structure of the prefusion form of measles virus fusion2 protein in complex with a fusion inhibitor compound (as-48)
38 d2ewla1
not modelled
23.0
14
Fold: E7 C-terminal domain-likeSuperfamily: E7 C-terminal domain-likeFamily: E7 C-terminal domain-like
39 c1ixrB_
not modelled
20.2
15
PDB header: hydrolaseChain: B: PDB Molecule: holliday junction dna helicase ruva;PDBTitle: ruva-ruvb complex
40 c3j6vJ_
not modelled
18.7
16
PDB header: ribosomeChain: J: PDB Molecule: 28s ribosomal protein s10, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
41 c2v3mF_
not modelled
18.7
29
PDB header: ribosomal proteinChain: F: PDB Molecule: naf1;PDBTitle: structure of the gar1 domain of naf1
42 c4ql6A_
not modelled
18.7
17
PDB header: hydrolaseChain: A: PDB Molecule: carboxy-terminal processing protease;PDBTitle: structure of c. trachomatis ct441
43 c4z7lB_
not modelled
17.6
29
PDB header: hydrolase/rnaChain: B: PDB Molecule: cas6b;PDBTitle: crystal structure of cas6b
44 c4w8xA_
not modelled
15.8
14
PDB header: rna binding proteinChain: A: PDB Molecule: crispr system cmr subunit cmr1-1;PDBTitle: crystal structure of cmr1 from pyrococcus furiosus bound to a2 nucleotide
45 d2aw2a1
not modelled
14.6
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)
46 c3a14B_
not modelled
12.6
12
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
47 c1hjpA_
not modelled
12.4
11
PDB header: dna recombinationChain: A: PDB Molecule: ruva;PDBTitle: holliday junction binding protein ruva from e. coli
48 c3mgjA_
not modelled
12.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mj1480;PDBTitle: crystal structure of the saccharop_dh_n domain of mj1480 protein from2 methanococcus jannaschii. northeast structural genomics consortium3 target mjr83a.
49 d1xaua_
not modelled
11.8
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains
50 c1k5hB_
not modelled
10.3
16
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose-5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose-5-phosphate reductoisomerase
51 c2ovsB_
not modelled
9.3
14
PDB header: gene regulation, ligand binding proteinChain: B: PDB Molecule: l0044;PDBTitle: crystal strcuture of a type three secretion system protein
52 c5hkqI_
not modelled
8.9
16
PDB header: toxin/antitoxinChain: I: PDB Molecule: cdii immunity protein;PDBTitle: crystal structure of cdi complex from escherichia coli stec_o31
53 d2gycf2
not modelled
8.9
39
Fold: MbtH/L9 domain-likeSuperfamily: L9 N-domain-likeFamily: Ribosomal protein L9 N-domain
54 d1cqua_
not modelled
8.4
39
Fold: MbtH/L9 domain-likeSuperfamily: L9 N-domain-likeFamily: Ribosomal protein L9 N-domain
55 d1f06a2
not modelled
7.1
28
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
56 c3gqqD_
not modelled
6.8
15
PDB header: splicingChain: D: PDB Molecule: protein unc-119 homolog a;PDBTitle: crystal structure of the human retinal protein 4 (unc-1192 homolog a). northeast structural genomics consortium3 target hr3066a
57 c1ixrA_
not modelled
6.8
28
PDB header: hydrolaseChain: A: PDB Molecule: holliday junction dna helicase ruva;PDBTitle: ruva-ruvb complex
58 c6iqsD_
not modelled
6.6
11
PDB header: hydrolaseChain: D: PDB Molecule: tail-specific protease;PDBTitle: crystal structure of prc with l245a and l340g mutations in complex2 with nlpi
59 c1v1cA_
not modelled
6.5
47
PDB header: sh3-domainChain: A: PDB Molecule: obscurin;PDBTitle: solution structure of the sh3 domain of obscurin
60 c5mjrA_
not modelled
6.3
50
PDB header: photosynthesisChain: A: PDB Molecule: protein thf1;PDBTitle: structure of psb29 at 1.55a
61 d1k4na_
not modelled
6.3
8
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Hypothetical protein YecM (EC4020)
62 c1pnyF_
not modelled
6.2
39
PDB header: ribosomeChain: F: PDB Molecule: 50s ribosomal protein l9;PDBTitle: crystal structure of the wild type ribosome from e. coli, 50s subunit2 of 70s ribosome. this file, 1pny, contains only molecules of the 50s3 ribosomal subunit. the 30s subunit is in the pdb file 1pnx.
63 c3nvnA_
not modelled
6.1
18
PDB header: viral protein/signaling proteinChain: A: PDB Molecule: evm139;PDBTitle: molecular mechanism of guidance cue recognition
64 d1nf1a_
not modelled
5.8
18
Fold: GTPase activation domain, GAPSuperfamily: GTPase activation domain, GAPFamily: p120GAP domain-like
65 c4oe4A_
not modelled
5.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: delta-1-pyrroline-5-carboxylate dehydrogenase,PDBTitle: crystal structure of yeast aldh4a1 complexed with nad+
66 c5wqlC_
not modelled
5.2
11
PDB header: protein binding/signaling protein/hydrolChain: C: PDB Molecule: tail-specific protease;PDBTitle: structure of a pdz-protease bound to a substrate-binding adaptor
67 d1b74a1
not modelled
5.2
50
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase
68 d1bf4a_
not modelled
5.2
11
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea
69 c2eqnA_
not modelled
5.0
29
PDB header: transcriptionChain: A: PDB Molecule: hypothetical protein loc92345;PDBTitle: solution structure of the naf1 domain of hypothetical2 protein bc008207 [homo sapiens]