| 1 |
|
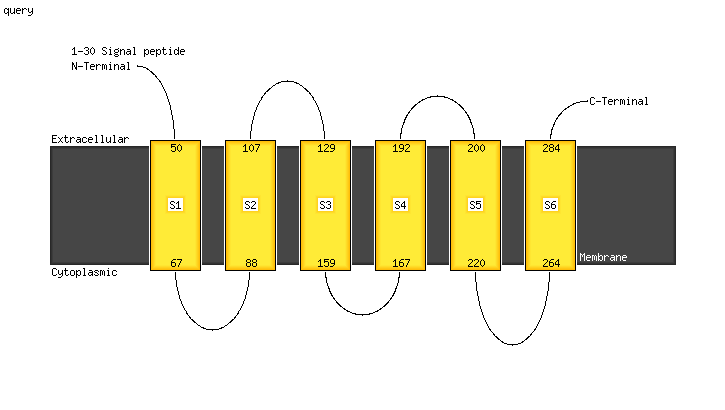
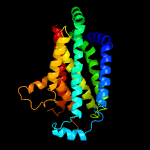
PDB 6an7 chain C
Region: 29 - 288
Aligned: 239
Modelled: 260
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: C: PDB Molecule:transport permease protein;
PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
Phyre2
| 2 |
|
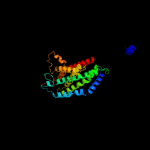
PDB 6an7 chain D
Region: 29 - 288
Aligned: 237
Modelled: 260
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: D: PDB Molecule:transport permease protein;
PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
Phyre2
| 3 |
|
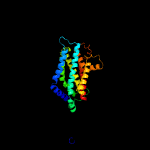
PDB 5njg chain B
Region: 27 - 288
Aligned: 253
Modelled: 262
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: B: PDB Molecule:atp-binding cassette sub-family g member 2;
PDBTitle: structure of an abc transporter: part of the structure that could be2 built de novo
Phyre2
| 4 |
|
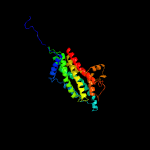
PDB 5nj3 chain B
Region: 8 - 288
Aligned: 271
Modelled: 281
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: B: PDB Molecule:atp-binding cassette sub-family g member 2;
PDBTitle: structure of an abc transporter: complete structure
Phyre2
| 5 |
|
PDB 5do7 chain B
Region: 22 - 288
Aligned: 258
Modelled: 267
Confidence: 100.0%
Identity: 9%
PDB header:transport protein
Chain: B: PDB Molecule:atp-binding cassette sub-family g member 8;
PDBTitle: crystal structure of the human sterol transporter abcg5/abcg8
Phyre2
| 6 |
|
PDB 5do7 chain A
Region: 12 - 288
Aligned: 269
Modelled: 277
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:atp-binding cassette sub-family g member 5;
PDBTitle: crystal structure of the human sterol transporter abcg5/abcg8
Phyre2
| 7 |
|
PDB 5xjy chain A
Region: 80 - 289
Aligned: 201
Modelled: 210
Confidence: 99.2%
Identity: 6%
PDB header:transport protein
Chain: A: PDB Molecule:atp-binding cassette sub-family a member 1;
PDBTitle: cryo-em structure of human abca1
Phyre2
| 8 |
|
PDB 5gas chain N
Region: 46 - 235
Aligned: 190
Modelled: 190
Confidence: 45.1%
Identity: 10%
PDB header:hydrolase
Chain: N: PDB Molecule:archaeal/vacuolar-type h+-atpase subunit i;
PDBTitle: thermus thermophilus v/a-atpase, conformation 2
Phyre2
| 9 |
|
PDB 5ir6 chain B
Region: 31 - 287
Aligned: 225
Modelled: 237
Confidence: 12.1%
Identity: 8%
PDB header:oxidoreductase
Chain: B: PDB Molecule:bd-type quinol oxidase subunit ii;
PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
Phyre2
| 10 |
|
PDB 3cqy chain A
Region: 1 - 8
Aligned: 8
Modelled: 2
Confidence: 11.4%
Identity: 38%
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of a functionally unknown protein (so_1313) from2 shewanella oneidensis mr-1
Phyre2
| 11 |
|
PDB 4yh5 chain B
Region: 1 - 7
Aligned: 7
Modelled: 7
Confidence: 10.2%
Identity: 43%
PDB header:transferase
Chain: B: PDB Molecule:levoglucosan kinase;
PDBTitle: lipomyces starkeyi levoglucosan kinase bound to adp and manganese
Phyre2
| 12 |
|
PDB 5n9y chain B
Region: 222 - 288
Aligned: 64
Modelled: 67
Confidence: 9.7%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:zinc transport protein zntb;
PDBTitle: the full-length structure of zntb
Phyre2
| 13 |
|
PDB 3rko chain F
Region: 37 - 117
Aligned: 81
Modelled: 81
Confidence: 8.4%
Identity: 11%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 14 |
|
PDB 2k9y chain A
Region: 251 - 288
Aligned: 38
Modelled: 38
Confidence: 7.9%
Identity: 13%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 15 |
|
PDB 2k9y chain B
Region: 251 - 288
Aligned: 38
Modelled: 38
Confidence: 7.9%
Identity: 13%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 16 |
|
PDB 2x7l chain P
Region: 8 - 40
Aligned: 33
Modelled: 33
Confidence: 7.3%
Identity: 21%
PDB header:immune system
Chain: P: PDB Molecule:protein rev;
PDBTitle: implications of the hiv-1 rev dimer structure at 3.2a resolution for2 multimeric binding to the rev response element
Phyre2
| 17 |
|
PDB 3lph chain D
Region: 8 - 40
Aligned: 33
Modelled: 33
Confidence: 6.1%
Identity: 21%
PDB header:viral protein
Chain: D: PDB Molecule:protein rev;
PDBTitle: crystal structure of the hiv-1 rev dimer
Phyre2
| 18 |
|
PDB 5ir6 chain A
Region: 79 - 288
Aligned: 203
Modelled: 210
Confidence: 5.1%
Identity: 16%
PDB header:oxidoreductase
Chain: A: PDB Molecule:bd-type quinol oxidase subunit i;
PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
Phyre2