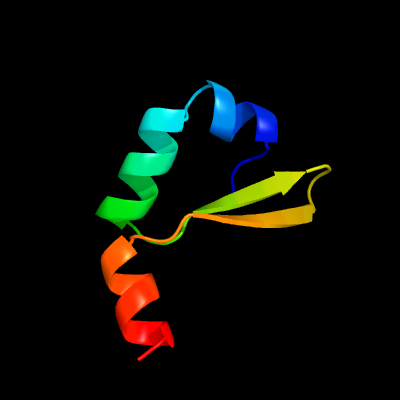
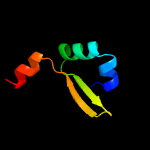
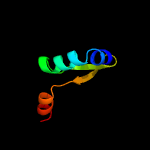
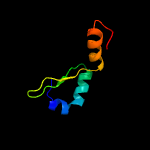
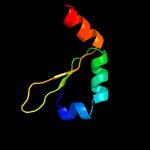
1 c2odkD_
99.1
23
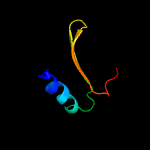
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
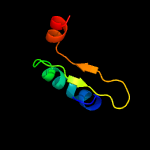
2 d2odka1
99.1
24
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like
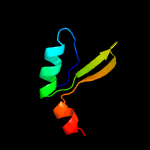
3 c3hryA_
98.7
23
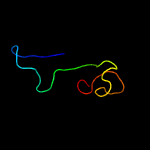
PDB header: antitoxinChain: A: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd in a trigonal space group and partially2 disordered
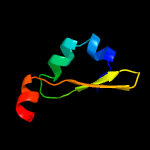
4 c3hs2H_
98.7
27
PDB header: antitoxinChain: H: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd truncated to residue 57 in an orthorhombic2 space group
5 d2a6qb1
98.2
26
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like
6 c3g5oA_
98.1
22
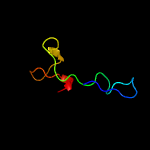
PDB header: toxin/antitoxinChain: A: PDB Molecule: uncharacterized protein rv2865;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
7 d2a6qa1
97.8
26
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like
8 c3d55A_
97.6
26
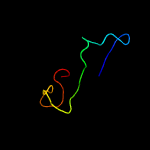
PDB header: toxin inhibitorChain: A: PDB Molecule: uncharacterized protein rv3357/mt3465;PDBTitle: crystal structure of m. tuberculosis yefm antitoxin
9 c3oeiB_
97.6
26
PDB header: toxin, protein bindingChain: B: PDB Molecule: relj (antitoxin rv3357);PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
10 c3k6qB_
78.0
14
PDB header: ligand binding proteinChain: B: PDB Molecule: putative ligand binding protein;PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
11 c1skoA_
68.3
22
PDB header: signaling proteinChain: A: PDB Molecule: mitogen-activated protein kinase kinase 1PDBTitle: mp1-p14 complex
12 d3cpta1
58.2
21
Fold: Profilin-likeSuperfamily: Roadblock/LC7 domainFamily: Roadblock/LC7 domain
13 d1p2za2
37.0
35
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group II dsDNA viruses VPFamily: Adenovirus hexon
14 c2bviK_
26.4
35
PDB header: virusChain: K: PDB Molecule: hexon protein;PDBTitle: the quasi-atomic model of human adenovirus type 52 capsid (part 2)
15 c3zifD_
26.2
35
PDB header: virusChain: D: PDB Molecule: hexon protein;PDBTitle: cryo-em structures of two intermediates provide insight into2 adenovirus assembly and disassembly
16 d2ns0a1
24.8
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like
17 d1ogda_
24.3
30
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like
18 c2wcvI_
22.0
19
PDB header: isomeraseChain: I: PDB Molecule: l-fucose mutarotase;PDBTitle: crystal structure of bacterial fucu
19 c4zdtB_
21.6
27
PDB header: hydrolaseChain: B: PDB Molecule: structure-specific endonuclease subunit slx4;PDBTitle: crystal structure of the ring finger domain of slx1 in complex with2 the c-terminal domain of slx4
20 c3e7nB_
21.2
26
PDB header: transport proteinChain: B: PDB Molecule: d-ribose high-affinity transport system;PDBTitle: crystal structure of d-ribose high-affinity transport system from2 salmonella typhimurium lt2
21 c3p13B_
not modelled
20.0
26
PDB header: isomeraseChain: B: PDB Molecule: d-ribose pyranase;PDBTitle: complex structure of d-ribose pyranase sa240 with d-ribose
22 c2wcuB_
not modelled
18.9
30
PDB header: isomeraseChain: B: PDB Molecule: protein fucu homolog;PDBTitle: crystal structure of mammalian fucu
23 c4a34L_
not modelled
18.1
23
PDB header: isomeraseChain: L: PDB Molecule: rbsd/fucu transport protein family protein;PDBTitle: crystal structure of the fucose mutarotase in complex with2 l-fucose from streptococcus pneumoniae
24 c3mvkA_
not modelled
16.9
23
PDB header: isomeraseChain: A: PDB Molecule: protein fucu;PDBTitle: the crystal structure of fucu from bifidobacterium longum to 1.65a
25 c4v0bA_
not modelled
15.7
19
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent zinc metalloprotease ftsh;PDBTitle: escherichia coli ftsh hexameric n-domain
26 d2ob5a1
not modelled
15.7
27
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like
27 d2ve8a1
not modelled
14.6
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like
28 c2muyA_
not modelled
14.4
15
PDB header: nucleotide binding proteinChain: A: PDB Molecule: atp-dependent zinc metalloprotease ftsh;PDBTitle: the solution structure of the ftsh periplasmic n-domain
29 d1x6va1
not modelled
14.0
15
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ATP sulfurylase N-terminal domain
30 d2j5pa1
not modelled
13.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FtsK C-terminal domain-like
31 c5ax7A_
not modelled
12.8
27
PDB header: transferaseChain: A: PDB Molecule: pyruvyl transferase 1;PDBTitle: yeast pyruvyltransferase pvg1p
32 c2inyA_
not modelled
12.3
33
PDB header: viral proteinChain: A: PDB Molecule: hexon protein;PDBTitle: nanoporous crystals of chicken embryo lethal orphan (celo) adenovirus2 major coat protein, hexon
33 d1p94a_
not modelled
12.2
27
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
34 c1gk7A_
not modelled
11.0
24
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 1a fragment (1a)
35 c3tuoA_
not modelled
10.8
25
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein satb1;PDBTitle: crystal structure of n-terminal domain of dna-binding protein satb1
36 c4heoA_
not modelled
10.8
50
PDB header: viral proteinChain: A: PDB Molecule: phosphoprotein;PDBTitle: hendra virus phosphoprotein c terminal domain
37 c4jj0B_
not modelled
10.7
10
PDB header: electron transportChain: B: PDB Molecule: mamp;PDBTitle: crystal structure of mamp
38 d2hq7a1
not modelled
10.6
9
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like
39 c6f9gE_
not modelled
10.4
16
PDB header: signaling proteinChain: E: PDB Molecule: methyl-accepting chemotaxis protein mcpu;PDBTitle: ligand binding domain of p. putida kt2440 polyamine chemorecpetors2 mcpu in complex putrescine.
40 c2vm2C_
not modelled
9.7
10
PDB header: oxidoreductaseChain: C: PDB Molecule: thioredoxin h isoform 1.;PDBTitle: crystal structure of barley thioredoxin h isoform 1 crystallized using2 peg as precipitant
41 c5flvA_
not modelled
9.1
22
PDB header: transcriptionChain: A: PDB Molecule: homeobox protein nkx-2.5, t-box transcription factor tbx5;PDBTitle: crystal structure of nkx2-5 and tbx5 bound to the nppa2 promoter region
42 c5lddA_
not modelled
8.7
30
PDB header: protein transportChain: A: PDB Molecule: mon1;PDBTitle: crystal structure of the heterodimeric gef mon1-ccz1 in complex with2 ypt7
43 d1y8xb1
not modelled
8.5
25
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)
44 c5yadA_
not modelled
8.4
25
PDB header: rna binding proteinChain: A: PDB Molecule: meiosis regulator and mrna stability factor 1;PDBTitle: crystal structure of marf1 lotus domain from mus musculus
45 d1f8fa2
not modelled
8.3
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
46 c5zf2A_
not modelled
8.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin (h-type,trx-h);PDBTitle: crystal structure of trxlp from edwardsiella tarda eib202
47 c3v62C_
not modelled
8.0
50
PDB header: protein binding/dna binding proteinChain: C: PDB Molecule: atp-dependent dna helicase srs2;PDBTitle: structure of the s. cerevisiae srs2 c-terminal domain in complex with2 pcna conjugated to sumo on lysine 164
48 c3v62F_
not modelled
8.0
50
PDB header: protein binding/dna binding proteinChain: F: PDB Molecule: atp-dependent dna helicase srs2;PDBTitle: structure of the s. cerevisiae srs2 c-terminal domain in complex with2 pcna conjugated to sumo on lysine 164
49 c5u1sA_
not modelled
8.0
22
PDB header: hydrolaseChain: A: PDB Molecule: separin;PDBTitle: crystal structure of the saccharomyces cerevisiae separase-securin2 complex at 3.0 angstrom resolution
50 c4cgbE_
not modelled
7.9
37
PDB header: cell cycleChain: E: PDB Molecule: echinoderm microtubule-associated protein-like 2;PDBTitle: crystal structure of the trimerization domain of eml2
51 c3s4rB_
not modelled
7.7
24
PDB header: structural proteinChain: B: PDB Molecule: vimentin;PDBTitle: crystal structure of vimentin coil1a/1b fragment with a stabilizing2 mutation
52 c1u2zC_
not modelled
7.5
18
PDB header: transferaseChain: C: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-79PDBTitle: crystal structure of histone k79 methyltransferase dot1p2 from yeast
53 d1u2za_
not modelled
7.3
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Catalytic, N-terminal domain of histone methyltransferase Dot1l
54 c3ssmB_
not modelled
7.0
25
PDB header: transferaseChain: B: PDB Molecule: methyltransferase;PDBTitle: myce methyltransferase from the mycinamycin biosynthetic pathway in2 complex with mg and sah, crystal form 1
55 c4cgbA_
not modelled
6.8
37
PDB header: cell cycleChain: A: PDB Molecule: echinoderm microtubule-associated protein-like 2;PDBTitle: crystal structure of the trimerization domain of eml2
56 c2nr1A_
not modelled
6.8
33
PDB header: receptorChain: A: PDB Molecule: nr1 m2;PDBTitle: transmembrane segment 2 of nmda receptor nr1, nmr, 102 structures
57 c5jieE_
not modelled
6.7
18
PDB header: viral proteinChain: E: PDB Molecule: protein delta;PDBTitle: crystal structure of the orsay virus delta protein n-terminal fragment2 (aa 1~66)
58 c3louB_
not modelled
6.7
17
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
59 c4euyA_
not modelled
6.5
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of thioredoxin-like protein bce_0499 from bacillus2 cereus atcc 10987
60 c1b9xC_
not modelled
6.4
16
PDB header: signaling proteinChain: C: PDB Molecule: protein (phosducin);PDBTitle: structural analysis of phosducin and its phosphorylation-2 regulated interaction with transducin
61 c2htfA_
not modelled
6.4
33
PDB header: transferaseChain: A: PDB Molecule: dna polymerase mu;PDBTitle: the solution structure of the brct domain from human2 polymerase reveals homology with the tdt brct domain
62 d1g8fa1
not modelled
6.3
19
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ATP sulfurylase N-terminal domain
63 c2qsiB_
not modelled
6.3
3
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative hydrogenase expression/formation protein hupg;PDBTitle: crystal structure of putative hydrogenase expression/formation protein2 hupg from rhodopseudomonas palustris cga009
64 c2wtoB_
not modelled
6.2
17
PDB header: metal binding proteinChain: B: PDB Molecule: orf131 protein;PDBTitle: crystal structure of apo-form czce from c. metallidurans ch34
65 d1t3ta7
not modelled
6.1
25
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like
66 c2jvzA_
not modelled
6.1
15
PDB header: splicingChain: A: PDB Molecule: far upstream element-binding protein 2;PDBTitle: solution nmr structure of the second and third kh domains2 of ksrp
67 c2p22D_
not modelled
6.0
10
PDB header: transport proteinChain: D: PDB Molecule: hypothetical 12.0 kda protein in ade3-ser2 intergenicPDBTitle: structure of the yeast escrt-i heterotetramer core
68 c4ewvB_
not modelled
6.0
22
PDB header: ligaseChain: B: PDB Molecule: 4-substituted benzoates-glutamate ligase gh3.12;PDBTitle: crystal structure of gh3.12 in complex with ampcpp
69 c5uc2C_
not modelled
5.9
24
PDB header: hydrolaseChain: C: PDB Molecule: domain of unknown function duf1849;PDBTitle: crystal structure of beta-barrel-like, putative atp binding protein of2 domain of unknown function duf1849 from brucella abortus
70 c2pptA_
not modelled
5.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin-2;PDBTitle: crystal structure of thioredoxin-2
71 d2ff4a1
not modelled
5.8
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like
72 d2g50a3
not modelled
5.8
13
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
73 c3f1iH_
not modelled
5.6
17
PDB header: protein bindingChain: H: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: human escrt-0 core complex
74 d1pkma1
not modelled
5.6
10
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain
75 c4q2jA_
not modelled
5.5
28
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein satb1;PDBTitle: a novel structure-based mechanism for dna-binding of satb1
76 d1umka1
not modelled
5.4
17
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
77 c4fxjB_
not modelled
5.4
13
PDB header: transferaseChain: B: PDB Molecule: pyruvate kinase isozymes m1/m2;PDBTitle: structure of m2 pyruvate kinase in complex with phenylalanine
78 d1qx4a1
not modelled
5.3
12
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
79 d2jhfa2
not modelled
5.3
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain
80 c1vjqB_
not modelled
5.3
41
PDB header: structural genomics, de novo proteinChain: B: PDB Molecule: designed protein;PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
81 c1v98A_
not modelled
5.2
24
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin;PDBTitle: crystal structure analysis of thioredoxin from thermus thermophilus
82 c5vf0B_
not modelled
5.1
24
PDB header: transferaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase rad18;PDBTitle: solution nmr structure of human rad18 (198-240) in complex with2 ubiquitin
83 c5w82E_
not modelled
5.0
19
PDB header: viral proteinChain: E: PDB Molecule: protein delta;PDBTitle: crystal structure of orsay virus delta protein n-terminal fragment (aa2 1-101)
84 c3o6uB_
not modelled
5.0
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein cpe2226;PDBTitle: crystal structure of cpe2226 protein from clostridium perfringens.2 northeast structural genomics consortium target cpr195