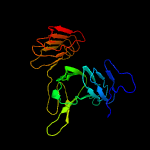
1 c1ygvA_
100.0
48
PDB header: structural protein/contractile proteinChain: A: PDB Molecule: collagen i alpha 1;PDBTitle: the structure of collagen type i. single type i collagen2 molecule: rigid refinment
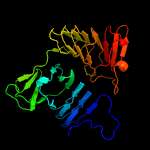
2 c3hqvB_
100.0
26
PDB header: structural protein, contractile proteinChain: B: PDB Molecule: collagen alpha-2(i) chain;PDBTitle: low resolution, molecular envelope structure of type i2 collagen in situ determined by fiber diffraction. single3 type i collagen molecule, rigid body refinement
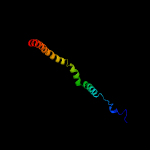
3 c1y0fB_
99.9
26
PDB header: structural protein/contractile proteinChain: B: PDB Molecule: collagen i alpha 2;PDBTitle: the structure of collagen type i. single type i collagen2 molecule
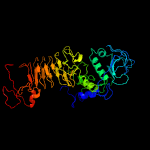
4 c5xfsA_
99.9
50
PDB header: protein transportChain: A: PDB Molecule: pe family protein pe8;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
5 d2g38a1
99.9
30
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PE
6 c2g38A_
99.9
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
7 c3bogB_
97.9
42
PDB header: antifreeze proteinChain: B: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
8 c3bogA_
97.9
42
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
9 c2pneA_
97.7
36
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: crystal structure of the snow flea antifreeze protein
10 c3boiB_
97.7
36
PDB header: antifreeze proteinChain: B: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein racemate
11 c3boiA_
97.7
36
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein racemate
12 c2qubG_
97.0
18
PDB header: hydrolaseChain: G: PDB Molecule: extracellular lipase;PDBTitle: crystal structure of extracellular lipase lipa from serratia2 marcescens
13 c1nayC_
96.7
25
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: gpp-foldon:x-ray structure
14 c2zj6A_
96.3
17
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of d337a mutant of pseudomonas sp. mis38 lipase
15 c3j04A_
95.9
10
PDB header: structural proteinChain: A: PDB Molecule: myosin-11;PDBTitle: em structure of the heavy meromyosin subfragment of chick smooth2 muscle myosin with regulatory light chain in phosphorylated state
16 c1k7qA_
95.5
12
PDB header: hydrolaseChain: A: PDB Molecule: secreted protease c;PDBTitle: prtc from erwinia chrysanthemi: e189a mutant
17 c2bkiA_
95.4
10
PDB header: motor protein/metal-binding proteinChain: A: PDB Molecule: unconventional myosin;PDBTitle: myosin vi nucleotide-free (mdinsert2-iq) crystal structure
18 c2ml3A_
95.4
16
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 6;PDBTitle: solution structure of alge6r3 subunit from the azotobacter vinelandii2 mannuronan c5-epimerase
19 c2ycuA_
95.2
7
PDB header: motor proteinChain: A: PDB Molecule: non muscle myosin 2c, alpha-actinin;PDBTitle: crystal structure of human non muscle myosin 2c in pre-power stroke2 state
20 c5ctiC_
95.2
22
PDB header: structural proteinChain: C: PDB Molecule: collagen alpha-1(i) chain,collagen alpha-3(ix) chain;PDBTitle: crystal structure of the type ix collagen nc2 hetero-trimerization2 domain with a guest fragment a2a1a1 of type i collagen (native form)
21 c5ctdB_
not modelled
94.9
26
PDB header: structural proteinChain: B: PDB Molecule: collagen alpha-2(i) chain,collagen alpha-2(ix) chain;PDBTitle: crystal structure of the type ix collagen nc2 hetero-trimerization2 domain with a guest fragment a2a1a1 of type i collagen
22 c1i84V_
not modelled
94.9
11
PDB header: contractile proteinChain: V: PDB Molecule: smooth muscle myosin heavy chain;PDBTitle: cryo-em structure of the heavy meromyosin subfragment of2 chicken gizzard smooth muscle myosin with regulatory light3 chain in the dephosphorylated state. only c alphas4 provided for regulatory light chain. only backbone atoms5 provided for s2 fragment.
23 c5ctdA_
not modelled
94.7
25
PDB header: structural proteinChain: A: PDB Molecule: collagen alpha-1(i) chain,collagen alpha-1(ix) chain;PDBTitle: crystal structure of the type ix collagen nc2 hetero-trimerization2 domain with a guest fragment a2a1a1 of type i collagen
24 c5tbyA_
not modelled
94.6
12
PDB header: contractile proteinChain: A: PDB Molecule: myosin-7;PDBTitle: human beta cardiac heavy meromyosin interacting-heads motif obtained2 by homology modeling (using swiss-model) of human sequence from3 aphonopelma homology model (pdb-3jbh), rigidly fitted to human beta-4 cardiac negatively stained thick filament 3d-reconstruction (emd-5 2240)
25 d1kapp1
not modelled
94.2
19
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
26 c2ml2A_
not modelled
93.5
20
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 6;PDBTitle: solution structure of alge6r2 subunit from the azotobacter vinelandii2 mannuronan c5-epimerase
27 c1satA_
not modelled
93.3
20
PDB header: hydrolase (serine protease)Chain: A: PDB Molecule: serratia protease;PDBTitle: crystal structure of the 50 kda metallo protease from s.2 marcescens
28 d1k7ia1
not modelled
93.1
19
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
29 d1sata1
not modelled
92.5
18
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
30 c2agmA_
not modelled
92.5
25
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: solution structure of the r-module from alge4
31 c1jiwP_
not modelled
92.1
26
PDB header: hydrolase/hyrolase inhibitorChain: P: PDB Molecule: alkaline metalloproteinase;PDBTitle: crystal structure of the apr-aprin complex
32 c1o1aP_
not modelled
92.1
29
PDB header: contractile proteinChain: P: PDB Molecule: skeletal muscle myosin ii;PDBTitle: molecular models of averaged rigor crossbridges from tomograms of2 insect flight muscle
33 d1g9ka1
not modelled
91.4
19
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
34 c4pd3B_
not modelled
90.2
5
PDB header: contractile proteinChain: B: PDB Molecule: nonmuscle myosin heavy chain b, alpha-actinin a chimeraPDBTitle: crystal structure of rigor-like human nonmuscle myosin-2b
35 c1om8A_
not modelled
89.8
25
PDB header: hydrolaseChain: A: PDB Molecule: serralysin;PDBTitle: crystal structure of a cold adapted alkaline protease from pseudomonas2 tac ii 18, co-crystallyzed with 10 mm edta
36 d1kk8a2
not modelled
89.4
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins
37 c2klwA_
not modelled
89.2
30
PDB header: de novo proteinChain: A: PDB Molecule: (pkg)10;PDBTitle: solution structure of an abc collagen heterotrimer reveals a2 single-register helix stabilized by electrostatic3 interactions
38 c5i0iB_
not modelled
88.3
17
PDB header: motor proteinChain: B: PDB Molecule: unconventional myosin-x;PDBTitle: crystal structure of myosin x motor domain with 2iq motifs in pre-2 powerstroke state
39 c2otgA_
not modelled
87.2
11
PDB header: contractile proteinChain: A: PDB Molecule: myosin heavy chain;PDBTitle: rigor-like structures of muscle myosins reveal key2 mechanical elements in the transduction pathways of this3 allosteric motor
40 c2cuoF_
not modelled
86.6
33
PDB header: structural proteinChain: F: PDB Molecule: collagen model peptide (pro-pro-gly)9;PDBTitle: collagen model peptide (pro-pro-gly)9
41 c2cuoC_
not modelled
86.6
33
PDB header: structural proteinChain: C: PDB Molecule: collagen model peptide (pro-pro-gly)9;PDBTitle: collagen model peptide (pro-pro-gly)9
42 c3dtpA_
not modelled
84.3
12
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth and cardiac muscle;PDBTitle: tarantula heavy meromyosin obtained by flexible docking to tarantula2 muscle thick filament cryo-em 3d-map
43 c1k6fC_
not modelled
84.2
31
PDB header: structural proteinChain: C: PDB Molecule: collagen triple helix;PDBTitle: crystal structure of the collagen triple helix model [(pro-pro-gly)2 10]3
44 c1k6fE_
not modelled
84.2
31
PDB header: structural proteinChain: E: PDB Molecule: collagen triple helix;PDBTitle: crystal structure of the collagen triple helix model [(pro-pro-gly)2 10]3
45 c1k6fF_
not modelled
84.2
31
PDB header: structural proteinChain: F: PDB Molecule: collagen triple helix;PDBTitle: crystal structure of the collagen triple helix model [(pro-pro-gly)2 10]3
46 c1k6fB_
not modelled
84.2
31
PDB header: structural proteinChain: B: PDB Molecule: collagen triple helix;PDBTitle: crystal structure of the collagen triple helix model [(pro-pro-gly)2 10]3
47 c1k6fA_
not modelled
84.2
31
PDB header: structural proteinChain: A: PDB Molecule: collagen triple helix;PDBTitle: crystal structure of the collagen triple helix model [(pro-pro-gly)2 10]3
48 c1k6fD_
not modelled
84.2
31
PDB header: structural proteinChain: D: PDB Molecule: collagen triple helix;PDBTitle: crystal structure of the collagen triple helix model [(pro-pro-gly)2 10]3
49 c2dfsA_
not modelled
83.6
19
PDB header: contractile protein/transport proteinChain: A: PDB Molecule: myosin-5a;PDBTitle: 3-d structure of myosin-v inhibited state
50 c3ah9A_
not modelled
82.4
33
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)9 at 1.1 a resolution
51 c2cuoB_
not modelled
81.9
31
PDB header: structural proteinChain: B: PDB Molecule: collagen model peptide (pro-pro-gly)9;PDBTitle: collagen model peptide (pro-pro-gly)9
52 c2cuoA_
not modelled
81.9
31
PDB header: structural proteinChain: A: PDB Molecule: collagen model peptide (pro-pro-gly)9;PDBTitle: collagen model peptide (pro-pro-gly)9
53 c2cuoD_
not modelled
81.9
31
PDB header: structural proteinChain: D: PDB Molecule: collagen model peptide (pro-pro-gly)9;PDBTitle: collagen model peptide (pro-pro-gly)9
54 c2cuoE_
not modelled
81.9
31
PDB header: structural proteinChain: E: PDB Molecule: collagen model peptide (pro-pro-gly)9;PDBTitle: collagen model peptide (pro-pro-gly)9
55 c3ah9D_
not modelled
81.2
36
PDB header: structural proteinChain: D: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)9 at 1.1 a resolution
56 c3ah9F_
not modelled
80.7
33
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)9 at 1.1 a resolution
57 c5cxlA_
not modelled
80.7
30
PDB header: toxinChain: A: PDB Molecule: bifunctional hemolysin/adenylate cyclase;PDBTitle: crystal structure of rtx domain block v of adenylate cyclase toxin2 from bordetella pertussis
58 c3ah9B_
not modelled
79.9
32
PDB header: structural proteinChain: B: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)9 at 1.1 a resolution
59 c3ah9C_
not modelled
79.9
32
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)9 at 1.1 a resolution
60 c3ah9E_
not modelled
79.9
32
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)9 at 1.1 a resolution
61 c2vb6A_
not modelled
79.3
12
PDB header: motor proteinChain: A: PDB Molecule: myosin vi;PDBTitle: myosin vi (md-insert2-cam, delta insert1) post-rigor state (2 crystal form 2)
62 c3w5mA_
not modelled
78.8
21
PDB header: hydrolaseChain: A: PDB Molecule: putative rhamnosidase;PDBTitle: crystal structure of streptomyces avermitilis alpha-l-rhamnosidase
63 c2bkhA_
not modelled
78.1
13
PDB header: motor protein/metal-binding proteinChain: A: PDB Molecule: unconventional myosin;PDBTitle: myosin vi nucleotide-free (mdinsert2) crystal structure
64 c4r8gE_
not modelled
76.9
21
PDB header: protein binding/calcium-binding proteinChain: E: PDB Molecule: unconventional myosin-ic;PDBTitle: crystal structure of myosin-1c tail in complex with calmodulin
65 c3p4gD_
not modelled
76.4
14
PDB header: antifreeze proteinChain: D: PDB Molecule: antifreeze protein;PDBTitle: x-ray crystal structure of a hyperactive, ca2+-dependent, beta-helical2 antifreeze protein from an antarctic bacterium
66 d2mysa2
not modelled
75.7
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins
67 c3lphD_
not modelled
75.5
65
PDB header: viral proteinChain: D: PDB Molecule: protein rev;PDBTitle: crystal structure of the hiv-1 rev dimer
68 c3a0mF_
not modelled
74.5
33
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
69 c3jbhA_
not modelled
73.8
14
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain striated muscle;PDBTitle: two heavy meromyosin interacting-heads motifs flexible docked into2 tarantula thick filament 3d-map allows in depth study of intra- and3 intermolecular interactions
70 c5uz7R_
not modelled
73.0
21
PDB header: signaling proteinChain: R: PDB Molecule: calcitonin receptor;PDBTitle: volta phase plate cryo-electron microscopy structure of a calcitonin2 receptor-heterotrimeric gs protein complex
71 c5szsC_
not modelled
71.6
21
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: glycan shield and epitope masking of a coronavirus spike protein2 observed by cryo-electron microscopy
72 c3j44h_
not modelled
71.4
53
PDB header: ribosome/protein transportChain: H: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: structure of the methanococcus jannaschii ribosome-secyebeta channel2 complex (50s ribosomal subunit)
73 c3j21h_
not modelled
71.4
53
PDB header: ribosomeChain: H: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
74 c5jbh0_
not modelled
71.0
53
PDB header: transcriptionChain: 0: PDB Molecule: 30s ribosomal protein el41;PDBTitle: cryo-em structure of a full archaeal ribosomal translation initiation2 complex in the p-in conformation
75 c5jb30_
not modelled
71.0
53
PDB header: translationChain: 0: PDB Molecule: 30s ribosomal protein el41;PDBTitle: cryo-em structure of a full archaeal ribosomal translation initiation2 complex in the p-remote conformation
76 c6ar3A_
not modelled
70.0
35
PDB header: rna binding protein/rna/dnaChain: A: PDB Molecule: gsi-iic rt;PDBTitle: structure of a thermostable group ii intron reverse transcriptase with2 template-primer and its functional and evolutionary implications3 (rt/duplex (se-met))
77 c3ob4A_
not modelled
69.1
17
PDB header: allergenChain: A: PDB Molecule: maltose abc transporter periplasmic protein, arah 2;PDBTitle: mbp-fusion protein of the major peanut allergen ara h 2
78 c5xxum_
not modelled
69.0
52
PDB header: ribosomeChain: M: PDB Molecule: ribosomal protein es12;PDBTitle: small subunit of toxoplasma gondii ribosome
79 c2x7lP_
not modelled
68.5
59
PDB header: immune systemChain: P: PDB Molecule: protein rev;PDBTitle: implications of the hiv-1 rev dimer structure at 3.2a resolution for2 multimeric binding to the rev response element
80 c6i9eE_
not modelled
66.7
42
PDB header: virusChain: E: PDB Molecule: major head protein;PDBTitle: thermophage p23-45 empty expanded capsid
81 c4dwcA_
not modelled
66.1
50
PDB header: viral proteinChain: A: PDB Molecule: e(rns) glycoprotein;PDBTitle: crystal structure of the glycoprotein erns from the pestivirus bvdv-12 in complex with zn ions
82 c5o5jO_
not modelled
64.5
30
PDB header: ribosomeChain: O: PDB Molecule: 30s ribosomal protein s15;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
83 d1g1xb_
not modelled
64.1
28
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
84 c4l79A_
not modelled
61.6
16
PDB header: motor protein/metal binding proteinChain: A: PDB Molecule: unconventional myosin-ib;PDBTitle: crystal structure of nucleotide-free myosin 1b residues 1-728 with2 bound calmodulin
85 c2lm4A_
not modelled
61.3
31
PDB header: protein bindingChain: A: PDB Molecule: succinate dehydrogenase assembly factor 2, mitochondrial;PDBTitle: solution nmr structure of mitochondrial succinate dehydrogenase2 assembly factor 2 from saccharomyces cerevisiae, northeast structural3 genomics consortium target yt682a
86 c3admC_
not modelled
60.9
35
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
87 c2d3fD_
not modelled
60.7
35
PDB header: structural proteinChain: D: PDB Molecule: collagen model peptides (pro-pro-gly)4-pro-hyp-PDBTitle: crystal structures of collagen model peptides (pro-pro-gly)2 4-pro-hyp-gly-(pro-pro-gly)4
88 c1x1kB_
not modelled
60.7
35
PDB header: structural proteinChain: B: PDB Molecule: host-guest peptide (pro-pro-gly)4-(pro-allohyp-PDBTitle: host-guest peptide (pro-pro-gly)4-(pro-allohyp-gly)-(pro-2 pro-gly)4
89 c2d3fE_
not modelled
60.7
35
PDB header: structural proteinChain: E: PDB Molecule: collagen model peptides (pro-pro-gly)4-pro-hyp-PDBTitle: crystal structures of collagen model peptides (pro-pro-gly)2 4-pro-hyp-gly-(pro-pro-gly)4
90 c1x1kD_
not modelled
60.7
35
PDB header: structural proteinChain: D: PDB Molecule: host-guest peptide (pro-pro-gly)4-(pro-allohyp-PDBTitle: host-guest peptide (pro-pro-gly)4-(pro-allohyp-gly)-(pro-2 pro-gly)4
91 c1x1kC_
not modelled
60.7
35
PDB header: structural proteinChain: C: PDB Molecule: host-guest peptide (pro-pro-gly)4-(pro-allohyp-PDBTitle: host-guest peptide (pro-pro-gly)4-(pro-allohyp-gly)-(pro-2 pro-gly)4
92 c1x1kA_
not modelled
60.7
35
PDB header: structural proteinChain: A: PDB Molecule: host-guest peptide (pro-pro-gly)4-(pro-allohyp-PDBTitle: host-guest peptide (pro-pro-gly)4-(pro-allohyp-gly)-(pro-2 pro-gly)4
93 c2d3fF_
not modelled
60.7
35
PDB header: structural proteinChain: F: PDB Molecule: collagen model peptides (pro-pro-gly)4-pro-hyp-PDBTitle: crystal structures of collagen model peptides (pro-pro-gly)2 4-pro-hyp-gly-(pro-pro-gly)4
94 d2a5yb2
not modelled
58.7
23
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD
95 c3nb0A_
not modelled
58.3
16
PDB header: transferaseChain: A: PDB Molecule: glycogen [starch] synthase isoform 2;PDBTitle: glucose-6-phosphate activated form of yeast glycogen synthase
96 c2ds2C_
not modelled
58.0
43
PDB header: plant proteinChain: C: PDB Molecule: sweet protein mabinlin-2 chain a;PDBTitle: crystal structure of mabinlin ii
97 c3admA_
not modelled
55.4
35
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
98 c3admF_
not modelled
55.4
35
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
99 c3admE_
not modelled
55.4
35
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
100 c3admB_
not modelled
55.4
35
PDB header: structural proteinChain: B: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-ser-gly-(pro-pro-gly)4
101 c5epmC_
not modelled
55.1
56
PDB header: toxin/immune systemChain: C: PDB Molecule: beta-theraphotoxin-cm1a;PDBTitle: ceratotoxin variant in complex with specific antibody fab fragment
102 c4clvB_
not modelled
54.8
9
PDB header: metal binding proteinChain: B: PDB Molecule: nickel-cobalt-cadmium resistance protein nccx;PDBTitle: crystal structure of dodecylphosphocholine-solubilized nccx2 from cupriavidus metallidurans 31a
103 c5b68A_
not modelled
54.3
16
PDB header: transferaseChain: A: PDB Molecule: 4-alpha-glucanotransferase;PDBTitle: crystal structure of apo amylomaltase from corynebacterium glutamicum
104 c3a08D_
not modelled
53.7
41
PDB header: structural proteinChain: D: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4, monoclinic, twinned crystal
105 c3a19F_
not modelled
53.7
41
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4_h monoclinic, twinned crystal
106 c2d3hD_
not modelled
53.7
41
PDB header: structural proteinChain: D: PDB Molecule: collagen model peptides (pro-pro-gly)4-hyp-hyp-PDBTitle: crystal structures of collagen model peptides (pro-pro-gly)2 4-hyp-hyp-gly-(pro-pro-gly)4
107 c3ulwA_
not modelled
53.3
9
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s15;PDBTitle: 30s ribosomal protein s15 from campylobacter jejuni
108 c3abnA_
not modelled
52.8
35
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: crystal structure of (pro-pro-gly)4-hyp-asp-gly-(pro-pro-gly)4 at 1.022 a
109 c5dg3D_
not modelled
51.9
19
PDB header: transferaseChain: D: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-PDBTitle: structure of pseudomonas aeruginosa lpxa in complex with udp-3-o-(r-3-2 hydroxydecanoyl)-glcnac
110 d1a32a_
not modelled
51.8
22
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
111 c2jr5A_
not modelled
50.9
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0350 protein vc_2471;PDBTitle: solution structure of upf0350 protein vc_2471. northeast structural2 genomics target vcr36
112 c3a0mA_
not modelled
49.2
32
PDB header: structural proteinChain: A: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
113 d1vs5o1
not modelled
48.7
22
Fold: S15/NS1 RNA-binding domainSuperfamily: S15/NS1 RNA-binding domainFamily: Ribosomal protein S15
114 c3a0mE_
not modelled
47.4
33
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-ovg-(ppg)4, monoclinic, twinned crystal
115 c2m1aA_
not modelled
47.3
52
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 rev arginine-rich motif (arm);PDBTitle: hiv-1 rev arm peptide (residues t34-r50)
116 c3skqA_
not modelled
47.3
13
PDB header: metal transportChain: A: PDB Molecule: mitochondrial distribution and morphology protein 38;PDBTitle: mdm38 is a 14-3-3-like receptor and associates with the protein2 synthesis machinery at the inner mitochondrial membrane
117 d1niya_
not modelled
46.8
44
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins
118 d1mb6a_
not modelled
46.6
56
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins
119 c1etfB_
not modelled
46.4
52
PDB header: viral protein/rnaChain: B: PDB Molecule: rev peptide;PDBTitle: rev response element (rre) rna complexed with rev peptide,2 nmr, minimized average structure
120 c1etgB_
not modelled
46.4
52
PDB header: viral protein/rnaChain: B: PDB Molecule: rev peptide;PDBTitle: rev response element (rre) rna complexed with rev peptide,2 nmr, 19 structures