| 1 |
|
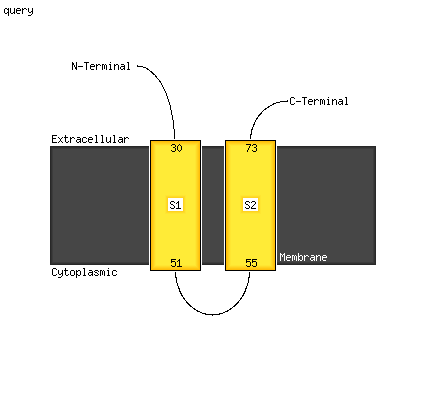
PDB 5xnm chain J
Region: 37 - 51
Aligned: 15
Modelled: 15
Confidence: 36.5%
Identity: 40%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
Phyre2
| 2 |
|
PDB 2i8g chain A domain 1
Region: 94 - 109
Aligned: 16
Modelled: 16
Confidence: 25.0%
Identity: 44%
Fold: Mog1p/PsbP-like
Superfamily: Mog1p/PsbP-like
Family: DIP2269-like
Phyre2
| 3 |
|
PDB 3jcu chain J
Region: 37 - 51
Aligned: 15
Modelled: 15
Confidence: 22.9%
Identity: 33%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
Phyre2
| 4 |
|
PDB 2axt chain J domain 1
Region: 37 - 51
Aligned: 15
Modelled: 15
Confidence: 21.0%
Identity: 40%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein J, PsbJ
Family: PsbJ-like
Phyre2
| 5 |
|
PDB 3a0h chain J
Region: 37 - 51
Aligned: 15
Modelled: 15
Confidence: 21.0%
Identity: 40%
PDB header:electron transport
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 6 |
|
PDB 4d6u chain I
Region: 82 - 98
Aligned: 17
Modelled: 17
Confidence: 13.2%
Identity: 53%
PDB header:oxidoreductase
Chain: I: PDB Molecule:cytochrome b-c1 complex subunit rieske, mitochondrial;
PDBTitle: cytochrome bc1 bound to the 4(1h)-pyridone gsk932121
Phyre2
| 7 |
|
PDB 2e76 chain G
Region: 41 - 58
Aligned: 18
Modelled: 18
Confidence: 9.2%
Identity: 61%
PDB header:photosynthesis
Chain: G: PDB Molecule:cytochrome b6-f complex subunit 5;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
Phyre2
| 8 |
|
PDB 1xop chain A
Region: 100 - 105
Aligned: 6
Modelled: 6
Confidence: 8.8%
Identity: 50%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: nmr structure of g1v mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
Phyre2
| 9 |
|
PDB 1xoo chain A
Region: 100 - 105
Aligned: 6
Modelled: 6
Confidence: 8.7%
Identity: 50%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: nmr structure of g1s mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
Phyre2
| 10 |
|
PDB 2yue chain A
Region: 80 - 110
Aligned: 31
Modelled: 31
Confidence: 8.6%
Identity: 32%
PDB header:rna binding protein
Chain: A: PDB Molecule:protein neuralized;
PDBTitle: solution structure of the neuz (nhr) domain in neuralized2 from drosophila melanogaster
Phyre2
| 11 |
|
PDB 5oeo chain C
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 8.0%
Identity: 50%
PDB header:membrane protein
Chain: C: PDB Molecule:transient receptor potential cation channel subfamily v
PDBTitle: solution structure of the complex of trpv5(655-725) with a calmodulin2 e32q/e68q double mutant
Phyre2
| 12 |
|
PDB 2jrd chain A
Region: 100 - 105
Aligned: 6
Modelled: 6
Confidence: 8.0%
Identity: 50%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: influenza hemagglutinin fusion domain mutant f9a
Phyre2
| 13 |
|
PDB 3rnv chain A
Region: 97 - 109
Aligned: 13
Modelled: 13
Confidence: 7.8%
Identity: 38%
PDB header:hydrolase
Chain: A: PDB Molecule:helper component proteinase;
PDBTitle: structure of the autocatalytic cysteine protease domain of potyvirus2 helper-component proteinase
Phyre2
| 14 |
|
PDB 2qm2 chain B
Region: 98 - 109
Aligned: 12
Modelled: 11
Confidence: 7.8%
Identity: 50%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative hopj type iii effector protein;
PDBTitle: putative hopj type iii effector protein from vibrio parahaemolyticus
Phyre2
| 15 |
|
PDB 4qh0 chain D
Region: 99 - 110
Aligned: 12
Modelled: 12
Confidence: 7.7%
Identity: 25%
PDB header:hydrolase
Chain: D: PDB Molecule:dna-entry nuclease (competence-specific nuclease);
PDBTitle: crystal structure of nuca from streptococcus agalactiae with magnesium2 ion bound
Phyre2
| 16 |
|
PDB 6n1b chain A
Region: 98 - 107
Aligned: 10
Modelled: 10
Confidence: 7.5%
Identity: 30%
PDB header:hydrolase
Chain: A: PDB Molecule:carbohydrate-binding protein;
PDBTitle: crystal structure of an n-acetylgalactosamine deacetylase from f.2 plautii in complex with blood group b trisaccharide
Phyre2
| 17 |
|
PDB 4zsf chain A
Region: 100 - 105
Aligned: 6
Modelled: 6
Confidence: 7.2%
Identity: 50%
PDB header:protein/dna
Chain: A: PDB Molecule:bsawi endonuclease;
PDBTitle: crystal structure of pre-specific restriction endonuclease bsawi-dna2 complex
Phyre2
| 18 |
|
PDB 1ibn chain A
Region: 100 - 105
Aligned: 6
Modelled: 6
Confidence: 7.1%
Identity: 50%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin ha2 chain peptide;
PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 5
Phyre2
| 19 |
|
PDB 1ibo chain A
Region: 100 - 105
Aligned: 6
Modelled: 6
Confidence: 7.1%
Identity: 50%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin ha2 chain peptide;
PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 7.4
Phyre2
| 20 |
|
PDB 2f3c chain I domain 1
Region: 80 - 85
Aligned: 6
Modelled: 6
Confidence: 6.8%
Identity: 83%
Fold: Kazal-type serine protease inhibitors
Superfamily: Kazal-type serine protease inhibitors
Family: Ovomucoid domain III-like
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|