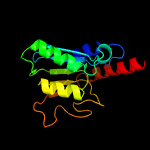
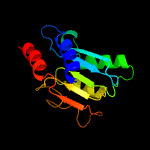
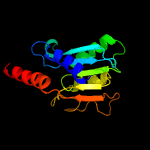
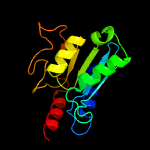
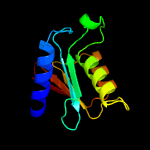
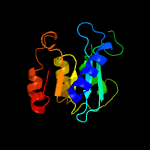
1 c2nx8A_
100.0
40
PDB header: hydrolaseChain: A: PDB Molecule: trna-specific adenosine deaminase;PDBTitle: the crystal structure of the trna-specific adenosine deaminase from2 streptococcus pyogenes
2 d2b3ja1
100.0
46
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
3 d1z3aa1
100.0
48
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
4 c3ocqA_
100.0
50
PDB header: hydrolaseChain: A: PDB Molecule: putative cytosine/adenosine deaminase;PDBTitle: crystal structure of trna-specific adenosine deaminase from salmonella2 enterica
5 d1wwra1
100.0
40
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
6 c3dh1D_
100.0
27
PDB header: hydrolaseChain: D: PDB Molecule: trna-specific adenosine deaminase 2;PDBTitle: crystal structure of human trna-specific adenosine-34 deaminase2 subunit adat2
7 d2g84a1
100.0
26
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
8 d1p6oa_
100.0
29
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
9 c5xkrA_
100.0
27
PDB header: hydrolaseChain: A: PDB Molecule: cmp/dcmp deaminase, zinc-binding protein;PDBTitle: crystal structure of msmeg3575 in complex with benzoguanamine
10 d2a8na1
100.0
44
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
11 d2b3za2
100.0
31
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
12 c2d5nB_
100.0
32
PDB header: hydrolase, oxidoreductaseChain: B: PDB Molecule: riboflavin biosynthesis protein ribd;PDBTitle: crystal structure of a bifunctional deaminase and reductase2 involved in riboflavin biosynthesis
13 c3zpgA_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: ribd;PDBTitle: acinetobacter baumannii ribd, form 2
14 c2o7pA_
100.0
33
PDB header: hydrolase, oxidoreductaseChain: A: PDB Molecule: riboflavin biosynthesis protein ribd;PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
15 c2hxvA_
100.0
31
PDB header: biosynthetic proteinChain: A: PDB Molecule: diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-PDBTitle: crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine2 deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828)3 from thermotoga maritima at 1.80 a resolution
16 d2hxva2
100.0
21
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
17 d1wkqa_
100.0
30
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
18 c2w4lC_
100.0
18
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidylate deaminase;PDBTitle: human dcmp deaminase
19 c5jfyC_
100.0
35
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidine deaminase;PDBTitle: crystal structure of a plant cytidine deaminase
20 c2hvwC_
100.0
23
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidylate deaminase;PDBTitle: crystal structure of dcmp deaminase from streptococcus mutans
21 c4p9eA_
not modelled
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: deoxycytidylate deaminase;PDBTitle: crystal structure of dcmp deaminase from the cyanophage s-tim5 in apo2 form
22 d1vq2a_
not modelled
100.0
29
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like
23 d1uwza_
not modelled
98.5
23
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
24 c3b8fB_
not modelled
98.5
14
PDB header: hydrolaseChain: B: PDB Molecule: putative blasticidin s deaminase;PDBTitle: crystal structure of the cytidine deaminase from bacillus anthracis
25 d1mq0a_
not modelled
98.5
18
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
26 d1r5ta_
not modelled
98.4
17
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
27 d2fr5a1
not modelled
98.4
18
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
28 d2d30a1
not modelled
98.4
20
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
29 c3dmoD_
not modelled
98.4
22
PDB header: hydrolaseChain: D: PDB Molecule: cytidine deaminase;PDBTitle: 1.6 a crystal structure of cytidine deaminase from burkholderia2 pseudomallei
30 c3ijfX_
not modelled
98.2
24
PDB header: hydrolaseChain: X: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium2 tuberculosis
31 c4eg2G_
not modelled
98.2
16
PDB header: hydrolaseChain: G: PDB Molecule: cytidine deaminase;PDBTitle: 2.2 angstrom crystal structure of cytidine deaminase from vibrio2 cholerae in complex with zinc and uridine
32 d1alna1
not modelled
98.2
27
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
33 c3r2nC_
not modelled
98.2
25
PDB header: hydrolaseChain: C: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase from mycobacterium leprae
34 d1alna2
not modelled
98.1
21
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
35 c1alnA_
not modelled
98.0
20
PDB header: hydrolaseChain: A: PDB Molecule: cytidine deaminase;PDBTitle: crystal structure of cytidine deaminase complexed with 3-deazacytidine
36 d2z3ga1
not modelled
97.8
17
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
37 c3oj6C_
not modelled
97.7
20
PDB header: hydrolaseChain: C: PDB Molecule: blasticidin-s deaminase;PDBTitle: crystal structure of blasticidin s deaminase from coccidioides immitis
38 c3vowB_
not modelled
95.5
19
PDB header: hydrolaseChain: B: PDB Molecule: probable dna dc->du-editing enzyme apobec-3c;PDBTitle: crystal structure of the human apobec3c having hiv-1 vif-binding2 interface
39 c3g8qA_
not modelled
95.1
34
PDB header: rna binding proteinChain: A: PDB Molecule: predicted rna-binding protein, contains thumpPDBTitle: a cytidine deaminase edits c-to-u in transfer rnas in2 archaea
40 c5tkmA_
not modelled
94.9
21
PDB header: hydrolaseChain: A: PDB Molecule: dna dc->du-editing enzyme apobec-3b;PDBTitle: crystal structure of human apobec3b n-terminal domain
41 c5k83C_
not modelled
94.8
19
PDB header: hydrolaseChain: C: PDB Molecule: apolipoprotein b mrna editing enzyme, catalytic peptide-PDBTitle: crystal structure of a primate apobec3g n-domain, in complex with2 ssdna
42 c6b0bE_
not modelled
92.3
21
PDB header: hydrolase/rnaChain: E: PDB Molecule: apobec3h;PDBTitle: crystal structure of human apobec3h
43 c2mzzA_
not modelled
91.9
21
PDB header: hydrolase, antiviral proteinChain: A: PDB Molecule: apolipoprotein b mrna-editing enzyme, catalyticPDBTitle: nmr structure of apobec3g ntd variant, sntd
44 c2kboA_
not modelled
90.8
19
PDB header: hydrolaseChain: A: PDB Molecule: dna dc->du-editing enzyme apobec-3g;PDBTitle: structure, interaction, and real-time monitoring of the2 enzymatic reaction of wild type apobec3g
45 c2nytB_
not modelled
88.1
26
PDB header: hydrolaseChain: B: PDB Molecule: probable c->u-editing enzyme apobec-2;PDBTitle: the apobec2 crystal structure and functional implications2 for aid
46 c6bwyA_
not modelled
85.9
22
PDB header: hydrolaseChain: A: PDB Molecule: protection of telomeres protein 1, dna dc->du-editingPDBTitle: dna substrate selection by apobec3g
47 c2m65A_
not modelled
82.9
26
PDB header: hydrolaseChain: A: PDB Molecule: probable dna dc->du-editing enzyme apobec-3a;PDBTitle: nmr structure of human restriction factor apobec3a
48 d2hi7b1
not modelled
28.5
43
Fold: Bromodomain-likeSuperfamily: DsbB-likeFamily: DsbB-like
49 c3e9jC_
not modelled
28.4
43
PDB header: oxidoreductaseChain: C: PDB Molecule: thiol/disulfide oxidoreductase dsbb;PDBTitle: structure of the charge-transfer intermediate of the2 transmembrane redox catalyst dsbb
50 c5mp0D_
not modelled
26.9
31
PDB header: hydrolaseChain: D: PDB Molecule: m7gpppn-mrna hydrolase;PDBTitle: human m7gpppn-mrna hydrolase (dcp2, nudt20) catalytic domain
51 c2k74A_
not modelled
25.9
29
PDB header: membrane protein, oxidoreductaseChain: A: PDB Molecule: disulfide bond formation protein b;PDBTitle: solution nmr structure of dsbb-ubiquinone complex
52 d2a6ta2
not modelled
23.7
25
Fold: NudixSuperfamily: NudixFamily: mRNA decapping enzyme-like
53 c4zbpC_
not modelled
22.9
13
PDB header: hydrolaseChain: C: PDB Molecule: nudix hydrolase 7;PDBTitle: crystal structure of the ampcpr-bound atnudt7
54 c2m1cA_
not modelled
20.4
13
PDB header: hydrolaseChain: A: PDB Molecule: dhh subfamily 1 protein;PDBTitle: haddock structure of gtyybt pas homodimer
55 d1xdpa4
not modelled
19.9
16
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain
56 d2o8ra4
not modelled
19.5
20
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain
57 c3l48B_
not modelled
19.1
36
PDB header: transport proteinChain: B: PDB Molecule: outer membrane usher protein papc;PDBTitle: crystal structure of the c-terminal domain of the papc usher
58 c3u3gA_
not modelled
18.2
23
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease h;PDBTitle: structure of lc11-rnase h1 isolated from compost by metagenomic2 approach: insight into the structural bases for unusual enzymatic3 properties of sto-rnase h1
59 d1xfna1
not modelled
14.6
33
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: PYP-like
60 c2xzn5_
not modelled
13.8
45
PDB header: ribosomeChain: 5: PDB Molecule: ribosomal protein s26e containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
61 c6bwsA_
not modelled
12.3
21
PDB header: unknown functionChain: A: PDB Molecule: glycolate utilization protein;PDBTitle: crystal structure of efga from methylobacterium extorquens
62 c3gxgA_
not modelled
12.0
15
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphatase (duf442);PDBTitle: crystal structure of putative phosphatase (duf442) (yp_001181608.1)2 from shewanella putrefaciens cn-32 at 1.60 a resolution
63 c3j38a_
not modelled
12.0
33
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein sa;PDBTitle: structure of the d. melanogaster 40s ribosomal proteins
64 c3ef5A_
not modelled
11.6
21
PDB header: hydrolaseChain: A: PDB Molecule: probable pyrophosphohydrolase;PDBTitle: structure of the rna pyrophosphohydrolase bdrpph in complex with dgtp
65 d1zcza2
not modelled
11.5
20
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC
66 d1jfua_
not modelled
11.4
26
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like
67 c6hiuA_
not modelled
10.8
27
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p460;PDBTitle: cytochrome p460 from methylococcus capsulatus (bath)
68 c2o8rA_
not modelled
10.7
20
PDB header: transferaseChain: A: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
69 c4nkpD_
not modelled
10.6
21
PDB header: chaperoneChain: D: PDB Molecule: putative extracellular heme-binding protein;PDBTitle: crystal structure of a putative extracellular heme-binding protein2 (despig_02683) from desulfovibrio piger atcc 29098 at 1.24 a3 resolution
70 c5xxua_
not modelled
10.4
40
PDB header: ribosomeChain: A: PDB Molecule: ribosomal protein us2;PDBTitle: small subunit of toxoplasma gondii ribosome
71 d1ttza_
not modelled
10.4
33
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase
72 c2xetB_
not modelled
9.9
17
PDB header: transport proteinChain: B: PDB Molecule: f1 capsule-anchoring protein;PDBTitle: conserved hydrophobic clusters on the surface of the caf1a usher2 c-terminal domain are important for f1 antigen assembly
73 c3u5ga_
not modelled
9.7
35
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein s0-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome b
74 c4a1oB_
not modelled
9.5
22
PDB header: transferase-hydrolaseChain: B: PDB Molecule: bifunctional purine biosynthesis protein purh;PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
75 d1j9ba_
not modelled
9.1
17
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: ArsC-like
76 c5xyia_
not modelled
9.1
35
PDB header: ribosomeChain: A: PDB Molecule: 40s ribosomal protein sa;PDBTitle: small subunit of trichomonas vaginalis ribosome
77 c1zy7A_
not modelled
8.9
33
PDB header: hydrolaseChain: A: PDB Molecule: rna-specific adenosine deaminase b1, isoformPDBTitle: crystal structure of the catalytic domain of an adenosine2 deaminase that acts on rna (hadar2) bound to inositol3 hexakisphosphate (ihp)
78 d1psqa_
not modelled
8.9
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like
79 d2a2la1
not modelled
8.9
23
Fold: Profilin-likeSuperfamily: GlcG-likeFamily: GlcG-like
80 d1vcda1
not modelled
8.5
38
Fold: NudixSuperfamily: NudixFamily: MutT-like
81 c6gyyB_
not modelled
8.4
27
PDB header: transferaseChain: B: PDB Molecule: diadenylate cyclase;PDBTitle: crystal structure of daca from staphylococcus aureus, n166c/t172c2 double mutant
82 c3zeyV_
not modelled
8.3
35
PDB header: ribosomeChain: V: PDB Molecule: ribosomal protein s26, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
83 c4pdeA_
not modelled
8.3
14
PDB header: hydrolaseChain: A: PDB Molecule: protein fdhd;PDBTitle: crystal structure of fdhd in complex with gdp
84 c2k1aA_
not modelled
8.1
57
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
85 c2kncA_
not modelled
8.1
50
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
86 d1nuia2
not modelled
7.9
27
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: DNA primase zinc finger
87 c4gpuA_
not modelled
7.7
13
PDB header: hydrolaseChain: A: PDB Molecule: klla0e02245p;PDBTitle: crystal structure of k. lactis dxo1 (ydr370c) in complex with2 manganese
88 c5y4uA_
not modelled
7.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-3;PDBTitle: crystal structure of grx domain of grx3 from saccharomyces cerevisiae
89 c3watA_
not modelled
7.3
12
PDB header: transferaseChain: A: PDB Molecule: 4-o-beta-d-mannosyl-d-glucose phosphorylase;PDBTitle: crystal structure of 4-o-beta-d-mannosyl-d-glucose phosphorylase mgp2 complexed with man+glc
90 c4ehiB_
not modelled
7.3
23
PDB header: hydrolase,transferaseChain: B: PDB Molecule: bifunctional purine biosynthesis protein purh;PDBTitle: an x-ray crystal structure of a putative bifunctional2 phosphoribosylaminoimidazolecarboxamide formyltransferase/imp3 cyclohydrolase
91 c2muqA_
not modelled
7.1
38
PDB header: ubiquitin binding proteinChain: A: PDB Molecule: fanconi anemia-associated protein of 20 kda;PDBTitle: solution structure of the human faap20 ubz
92 d1q98a_
not modelled
7.0
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like
93 c2murA_
not modelled
6.8
38
PDB header: protein bindingChain: A: PDB Molecule: fanconi anemia-associated protein of 20 kda;PDBTitle: solution structure of the human faap20 ubz-ubiquitin complex
94 c3a03A_
not modelled
6.7
22
PDB header: gene regulationChain: A: PDB Molecule: t-cell leukemia homeobox protein 2;PDBTitle: crystal structure of hox11l1 homeodomain
95 c2zvkU_
not modelled
6.5
38
PDB header: transferaseChain: U: PDB Molecule: dna polymerase eta;PDBTitle: crystal structure of pcna in complex with dna polymerase eta fragment
96 c6dknA_
not modelled
6.4
27
PDB header: hydrolaseChain: A: PDB Molecule: decapping nuclease dxo homolog, chloroplastic;PDBTitle: crystal structure of arabidopsis thaliana decapping nuclease dxo1
97 c5zvqA_
not modelled
6.4
18
PDB header: recombinationChain: A: PDB Molecule: recombination protein recr;PDBTitle: crystal structure of recombination mediator protein recr
98 c2yzhD_
not modelled
6.4
8
PDB header: oxidoreductaseChain: D: PDB Molecule: probable thiol peroxidase;PDBTitle: crystal structure of peroxiredoxin-like protein from aquifex aeolicus
99 c3ipzA_
not modelled
6.2
15
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-s14, chloroplastic;PDBTitle: crystal structure of arabidopsis monothiol glutaredoxin atgrxcp