1 c2ps3A_
100.0
19
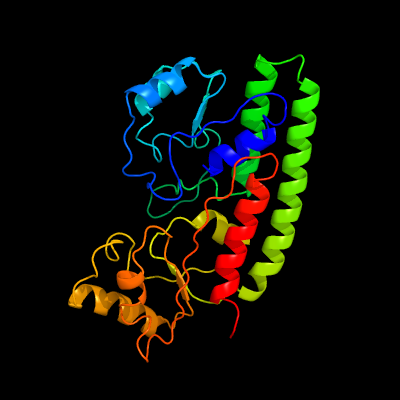
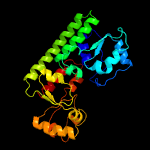
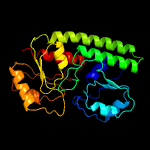
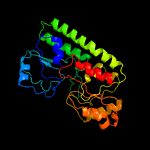
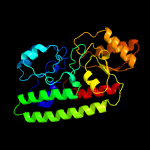
PDB header: metal transportChain: A: PDB Molecule: high-affinity zinc uptake system protein znua;PDBTitle: structure and metal binding properties of znua, a periplasmic zinc2 transporter from escherichia coli
2 c2ogwB_
100.0
18
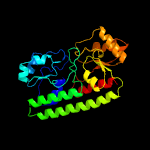
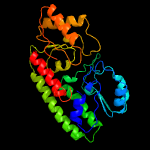
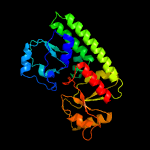
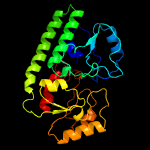
PDB header: transport proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znuaPDBTitle: structure of abc type zinc transporter from e. coli
3 c6r6kB_
100.0
22
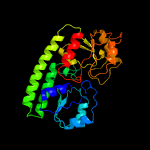
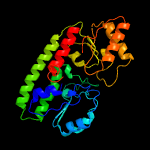
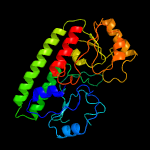
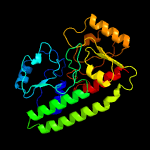
PDB header: protein transportChain: B: PDB Molecule: abc transporter substrate-binding protein;PDBTitle: structure of a fpvc mutant from pseudomonas aeruginosa
4 c2o1eB_
100.0
22
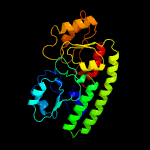
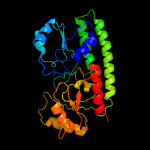
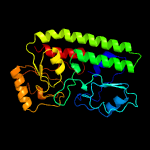
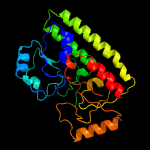
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ycdh;PDBTitle: crystal structure of the metal-dependent lipoprotein ycdh2 from bacillus subtilis, northeast structural genomics3 target sr583
5 c3hjtB_
100.0
21
PDB header: cell adhesion, transport proteinChain: B: PDB Molecule: lmb;PDBTitle: structure of laminin binding protein (lmb) of streptococcus agalactiae2 a bifunctional protein with adhesin and metal transporting activity
6 c4xrvB_
100.0
21
PDB header: metal binding proteinChain: B: PDB Molecule: periplasmic solute binding protein;PDBTitle: structure of a zn abc transporter substrate binding protein from2 paracoccus denitrificans
7 c5uyvA_
100.0
18
PDB header: metal transportChain: A: PDB Molecule: periplasmic chelated iron-binding protein yfea;PDBTitle: yfea ancillary sites that do not co-load with site 2
8 d1xvla1
100.0
24
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
9 d1toaa_
100.0
16
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
10 c1toaA_
100.0
16
PDB header: binding proteinChain: A: PDB Molecule: protein (periplasmic binding protein troa);PDBTitle: periplasmic zinc binding protein troa from treponema pallidum
11 c2ov3A_
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein component of an abc type zincPDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc bound
12 c4cl2A_
100.0
20
PDB header: transport proteinChain: A: PDB Molecule: periplasmic solute binding protein;PDBTitle: structure of periplasmic metal binding protein from candidatus2 liberibacter asiaticus
13 d1pq4a_
100.0
21
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
14 c5hx7A_
100.0
20
PDB header: transport proteinChain: A: PDB Molecule: manganese-binding lipoprotein mnta;PDBTitle: metal abc transporter from listeria monocytogenes
15 c3cx3A_
100.0
19
PDB header: metal binding proteinChain: A: PDB Molecule: lipoprotein;PDBTitle: crystal structure analysis of the streptococcus pneumoniae2 adcaii protein
16 c3mfqB_
100.0
16
PDB header: metal binding proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znua;PDBTitle: a glance into the metal binding specificity of troa: where elaborate2 behaviors occur in the active center
17 d1psza_
100.0
18
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
18 c4oxqB_
100.0
17
PDB header: metal binding proteinChain: B: PDB Molecule: manganese abc transporter, periplasmic-binding proteinPDBTitle: structure of staphylococcus pseudintermedius metal-binding protein2 sita in complex with zinc
19 c4k3vA_
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: abc superfamily atp binding cassette transporter, bindingPDBTitle: structure of staphylococcus aureus mntc
20 c1xvlC_
100.0
28
PDB header: metal transportChain: C: PDB Molecule: mn transporter;PDBTitle: the three-dimensional structure of mntc from synechocystis2 6803
21 c5jg7A_
not modelled
100.0
26
PDB header: metal transportChain: A: PDB Molecule: fur regulated salmonella iron transporter;PDBTitle: crystal structure of putative periplasmic binding protein from2 salmonella typhimurium lt2
22 c5n6yD_
not modelled
97.3
14
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase vanadium-iron protein alpha chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
23 d1qh8a_
not modelled
97.0
15
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
24 c3pdiB_
not modelled
96.6
18
PDB header: protein bindingChain: B: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nifn;PDBTitle: precursor bound nifen
25 d1m1na_
not modelled
96.4
9
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
26 c5kojD_
not modelled
96.3
15
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase femo beta subunit protein nifk;PDBTitle: nitrogenase mofep protein in the ids oxidized state
27 d1wo8a1
not modelled
96.3
19
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA
28 d2ay1a_
not modelled
96.3
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
29 d1mioa_
not modelled
96.2
10
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
30 c6f2cK_
not modelled
96.0
15
PDB header: lyaseChain: K: PDB Molecule: methylglyoxal synthase;PDBTitle: methylglyoxal synthase mgsa from bacillus subtilis
31 c2xdqB_
not modelled
95.7
14
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
32 d1m1nb_
not modelled
95.4
10
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
33 c3pdiG_
not modelled
95.3
14
PDB header: protein bindingChain: G: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nife;PDBTitle: precursor bound nifen
34 c4effA_
not modelled
95.3
14
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid aminotransferase;PDBTitle: crystal structure of aromatic-amino-acid aminotransferase from2 burkholderia pseudomallei
35 c3trjC_
not modelled
95.0
9
PDB header: isomeraseChain: C: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
36 c4rkdA_
not modelled
94.9
14
PDB header: transferaseChain: A: PDB Molecule: aromatic amino acid aminotransferase;PDBTitle: psychrophilic aromatic amino acids aminotransferase from psychrobacter2 sp. b6 cocrystalized with aspartic acid
37 c3aerB_
not modelled
94.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
38 c4wd2A_
not modelled
94.5
20
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid transaminase tyrb;PDBTitle: crystal structure of an aromatic amino acid aminotransferase from2 burkholderia cenocepacia j2315
39 d7aata_
not modelled
94.4
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
40 d1qh8b_
not modelled
94.4
12
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
41 c3mebB_
not modelled
94.2
12
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: structure of cytoplasmic aspartate aminotransferase from giardia2 lamblia
42 c2yvqA_
not modelled
94.0
17
PDB header: ligaseChain: A: PDB Molecule: carbamoyl-phosphate synthase;PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
43 d1vmda_
not modelled
94.0
20
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA
44 c5n6yE_
not modelled
93.9
14
PDB header: oxidoreductaseChain: E: PDB Molecule: nitrogenase vanadium-iron protein beta chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
45 c3h75A_
not modelled
93.9
6
PDB header: sugar binding proteinChain: A: PDB Molecule: periplasmic sugar-binding domain protein;PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
46 c3j21Z_
not modelled
93.6
15
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l30e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
47 c3zf7g_
not modelled
93.4
11
PDB header: ribosomeChain: G: PDB Molecule: PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
48 c5i01B_
not modelled
93.4
13
PDB header: isomeraseChain: B: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of phosphoheptose isomerase gmha from neisseria gonorrhoeae
49 c3tcmB_
not modelled
93.3
14
PDB header: transferaseChain: B: PDB Molecule: alanine aminotransferase 2;PDBTitle: crystal structure of alanine aminotransferase from hordeum vulgare
50 c3s99A_
not modelled
93.3
17
PDB header: lipid binding proteinChain: A: PDB Molecule: basic membrane lipoprotein;PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
51 d2csta_
not modelled
93.3
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
52 d1w41a1
not modelled
93.2
14
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: L30e/L7ae ribosomal proteins
53 c3cpqB_
not modelled
93.2
11
PDB header: ribosomal proteinChain: B: PDB Molecule: 50s ribosomal protein l30e;PDBTitle: crystal structure of l30e a ribosomal protein from2 methanocaldococcus jannaschii dsm2661 (mj1044)
54 d1t0kb_
not modelled
93.2
11
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: L30e/L7ae ribosomal proteins
55 c2zy4F_
not modelled
93.1
17
PDB header: lyaseChain: F: PDB Molecule: l-aspartate beta-decarboxylase;PDBTitle: dodecameric l-aspartate beta-decarboxylase
56 c4a1dG_
not modelled
93.1
16
PDB header: ribosomeChain: G: PDB Molecule: rpl30;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
57 d2bo1a1
not modelled
93.0
16
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: L30e/L7ae ribosomal proteins
58 d1yaaa_
not modelled
92.9
14
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
59 d1a9xa2
not modelled
92.8
19
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain
60 d1miob_
not modelled
92.7
12
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
61 d2ez9a1
not modelled
92.6
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
62 c5by2A_
not modelled
92.4
8
PDB header: isomeraseChain: A: PDB Molecule: phosphoheptose isomerase;PDBTitle: sedoheptulose 7-phosphate isomerase from colwellia psychrerythraea2 strain 34h
63 c6btmB_
not modelled
92.3
13
PDB header: membrane proteinChain: B: PDB Molecule: alternative complex iii subunit b;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
64 c4eu1A_
not modelled
92.2
10
PDB header: transferaseChain: A: PDB Molecule: mitochondrial aspartate aminotransferase;PDBTitle: structure of a mitochondrial aspartate aminotransferase from2 trypanosoma brucei
65 c6hnuA_
not modelled
92.2
12
PDB header: transferaseChain: A: PDB Molecule: aromatic amino acid aminotransferase i;PDBTitle: crystal structure of the aminotransferase aro8 from c. albicans with2 ligands
66 c4mlcA_
not modelled
92.0
17
PDB header: signaling proteinChain: A: PDB Molecule: extracellular ligand-binding receptor;PDBTitle: abc transporter substrate-binding protein fromdesulfitobacterium2 hafniense
67 c6dspB_
not modelled
92.0
18
PDB header: signaling proteinChain: B: PDB Molecule: autoinducer 2-binding protein lsrb;PDBTitle: lsrb from clostridium saccharobutylicum in complex with ai-2
68 c5braA_
not modelled
91.9
14
PDB header: solute-binding proteinChain: A: PDB Molecule: putative periplasmic binding protein with substrate ribose;PDBTitle: crystal structure of a putative periplasmic solute binding protein2 (ipr025997) from ochrobactrum anthropi atcc49188 (oant_2843, target3 efi-511085)
69 c3v7qB_
not modelled
91.7
11
PDB header: rna binding proteinChain: B: PDB Molecule: probable ribosomal protein ylxq;PDBTitle: crystal structure of b. subtilis ylxq at 1.55 a resolution
70 c3k7yA_
not modelled
91.5
11
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: aspartate aminotransferase of plasmodium falciparum
71 c2ynmD_
not modelled
91.4
15
PDB header: oxidoreductaseChain: D: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
72 c4rxuA_
not modelled
91.3
18
PDB header: transport proteinChain: A: PDB Molecule: periplasmic sugar-binding protein;PDBTitle: crystal structure of carbohydrate transporter solute binding protein2 caur_1924 from chloroflexus aurantiacus, target efi-511158, in3 complex with d-glucose
73 c4wzzA_
not modelled
91.3
15
PDB header: transport proteinChain: A: PDB Molecule: putative sugar abc transporter, substrate-binding protein;PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from clostridium phytofermentas (cphy_0583, target efi-3 511148) with bound l-rhamnose
74 c3zeyF_
not modelled
91.2
7
PDB header: ribosomeChain: F: PDB Molecule: 40s ribosomal protein s12;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
75 c4wwhA_
not modelled
91.2
14
PDB header: transport proteinChain: A: PDB Molecule: abc transporter;PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from mycobacterium smegmatis (msmeg_1704, target efi-3 510967) with bound d-galactose
76 d1ajsa_
not modelled
91.2
14
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
77 c2yvaB_
not modelled
91.0
14
PDB header: dna binding proteinChain: B: PDB Molecule: dnaa initiator-associating protein diaa;PDBTitle: crystal structure of escherichia coli diaa
78 d2ji7a1
not modelled
90.8
17
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
79 c3on1A_
not modelled
90.7
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh2414 protein;PDBTitle: the structure of a protein with unknown function from bacillus2 halodurans c
80 c4kzkA_
not modelled
90.7
15
PDB header: sugar binding proteinChain: A: PDB Molecule: l-arabinose abc transporter, periplasmic l-arabinose-PDBTitle: the structure of the periplasmic l-arabinose binding protein from2 burkholderia thailandensis
81 d2djia1
not modelled
90.6
17
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
82 c6ezlB_
not modelled
90.4
16
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of aspartate aminotransferase from trypanosoma cruzi2 at 2.07 angstrom resolution
83 c3g7qA_
not modelled
90.4
13
PDB header: transferaseChain: A: PDB Molecule: valine-pyruvate aminotransferase;PDBTitle: crystal structure of valine-pyruvate aminotransferase avta2 (np_462565.1) from salmonella typhimurium lt2 at 1.80 a resolution
84 d1ybha1
not modelled
89.9
19
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
85 c1ynuA_
not modelled
89.8
20
PDB header: lyaseChain: A: PDB Molecule: 1-aminocyclopropane-1-carboxylate synthase;PDBTitle: crystal structure of apple acc synthase in complex with l-vinylglycine
86 c6hndA_
not modelled
89.7
11
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid:2-oxoglutarate transaminase;PDBTitle: crystal structure of the aromatic aminotransferase aro9 from c.2 albicans
87 d3tata_
not modelled
89.7
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
88 c6c3aB_
not modelled
89.6
13
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: o2-, plp-dependent l-arginine hydroxylase rohp 4-hydroxy-2-2 ketoarginine complex
89 c2zkr6_
not modelled
89.4
15
PDB header: ribosomal protein/rnaChain: 6: PDB Molecule: 60s ribosomal protein l30e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
90 c4a0rB_
not modelled
89.2
19
PDB header: transferaseChain: B: PDB Molecule: adenosylmethionine-8-amino-7-oxononanoatePDBTitle: structure of bifunctional dapa aminotransferase-dtb synthetase from2 arabidopsis thaliana bound to dethiobiotin (dtb).
91 c3uugB_
not modelled
89.0
16
PDB header: sugar binding proteinChain: B: PDB Molecule: multiple sugar-binding periplasmic receptor chve;PDBTitle: crystal structure of the periplasmic sugar binding protein chve
92 c2lbwA_
not modelled
88.9
14
PDB header: rna binding proteinChain: A: PDB Molecule: h/aca ribonucleoprotein complex subunit 2;PDBTitle: solution structure of the s. cerevisiae h/aca rnp protein nhp2p-s82w2 mutant
93 d1iaya_
not modelled
88.7
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
94 c3f6tA_
not modelled
88.5
17
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of aspartate aminotransferase (e.c. 2.6.1.1)2 (yp_194538.1) from lactobacillus acidophilus ncfm at 2.15 a3 resolution
95 d1tjya_
not modelled
88.4
15
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like
96 c3jtxB_
not modelled
88.3
16
PDB header: transferaseChain: B: PDB Molecule: aminotransferase;PDBTitle: crystal structure of aminotransferase (np_283882.1) from neisseria2 meningitidis z2491 at 1.91 a resolution
97 d2q7wa1
not modelled
88.3
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
98 c1p1hD_
not modelled
88.2
22
PDB header: isomeraseChain: D: PDB Molecule: inositol-3-phosphate synthase;PDBTitle: crystal structure of the 1l-myo-inositol/nad+ complex
99 c2xznU_
not modelled
88.2
9
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein l7ae containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
100 c2zy3A_
not modelled
88.0
14
PDB header: lyaseChain: A: PDB Molecule: l-aspartate beta-decarboxylase;PDBTitle: dodecameric l-aspartate beta-decarboxylase
101 d1vkoa1
not modelled
88.0
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
102 c3nraA_
not modelled
87.9
17
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of an aspartate aminotransferase (yp_354942.1) from2 rhodobacter sphaeroides 2.4.1 at 2.15 a resolution
103 c3hmuA_
not modelled
87.8
18
PDB header: transferaseChain: A: PDB Molecule: aminotransferase, class iii;PDBTitle: crystal structure of a class iii aminotransferase from silicibacter2 pomeroyi
104 c4h51B_
not modelled
87.8
15
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of a putative aspartate aminotransferase from2 leishmania major friedlin
105 c4je5C_
not modelled
87.7
17
PDB header: transferaseChain: C: PDB Molecule: aromatic/aminoadipate aminotransferase 1;PDBTitle: crystal structure of the aromatic aminotransferase aro8, a putative2 alpha-aminoadipate aminotransferase in saccharomyces cerevisiae
106 d2byla1
not modelled
87.4
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
107 c4pz0A_
not modelled
87.3
9
PDB header: sugar binding proteinChain: A: PDB Molecule: sugar abc transporter, sugar-binding protein;PDBTitle: the crystal structure of a solute binding protein from bacillus2 anthracis str. ames in complex with quorum-sensing signal3 autoinducer-2 (ai-2)
108 c3o1hB_
not modelled
87.3
13
PDB header: signaling proteinChain: B: PDB Molecule: periplasmic protein tort;PDBTitle: crystal structure of the tors sensor domain - tort complex in the2 presence of tmao
109 c5ewrA_
not modelled
87.3
16
PDB header: rna binding proteinChain: A: PDB Molecule: box c/d snornp and u4 snrnp component snu13p;PDBTitle: c merolae u4 snrnp protein snu13
110 c5uowB_
not modelled
86.9
17
PDB header: membrane proteinChain: B: PDB Molecule: n-methyl-d-aspartate receptor subunit nr2a;PDBTitle: triheteromeric nmda receptor glun1/glun2a/glun2b in complex with2 glycine, glutamate, mk-801 and a glun2b-specific fab, at ph 6.5
111 c5xyiM_
not modelled
86.8
7
PDB header: ribosomeChain: M: PDB Molecule: ribosomal protein l7ae, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
112 c6irfD_
not modelled
86.6
19
PDB header: membrane proteinChain: D: PDB Molecule: glutamate receptor ionotropic, nmda 2a;PDBTitle: structure of the human glun1/glun2a nmda receptor in the2 glutamate/glycine-bound state at ph 6.3, class i
113 d1ozha1
not modelled
86.1
14
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
114 d2cc0a1
not modelled
86.0
11
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
115 c4y9tA_
not modelled
85.8
19
PDB header: solute-binding proteinChain: A: PDB Molecule: abc transporter, solute binding protein;PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from agrobacterium vitis s4 (avi_5305, target efi-511224)3 with bound alpha-d-glucosamine
116 c4pe6B_
not modelled
85.7
21
PDB header: solute-binding proteinChain: B: PDB Molecule: putative abc transporter;PDBTitle: crystal structure of abc transporter solute binding protein from2 thermobispora bispora dsm 43833
117 d1vqof1
not modelled
85.5
16
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: L30e/L7ae ribosomal proteins
118 c5xxuM_
not modelled
85.3
11
PDB header: ribosomeChain: M: PDB Molecule: ribosomal protein es12;PDBTitle: small subunit of toxoplasma gondii ribosome
119 c5c4nD_
not modelled
85.1
13
PDB header: oxidoreductaseChain: D: PDB Molecule: precorrin-6a reductase;PDBTitle: cobk precorrin-6a reductase
120 d1bw0a_
not modelled
85.0
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like